ISSN: 2206-7418Nanotheranostics
Nanotheranostics 2018; 2(3):271-279. doi:10.7150/ntno.24590 This issue Cite
Research Paper
Genome-wide Transcriptional Analysis of Oxidative Stress-related Genes and Pathways Induced by CdTe aqQDs in Mice
1. Department of Toxicology and Sanitary Chemistry, School of Public Health, Capital Medical University, Beijing, 100069, China
2. Beijing Key Laboratory of Environmental Toxicology, Capital Medical University, Beijing, 100069, China
3. Xue Yuan Road Community Health Service Centers, Beijing, 100069, China
†These authors contributed equally to this work
Abstract
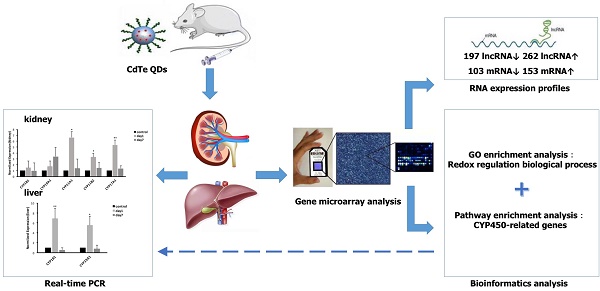
Objective: Quantum dots (QDs) has widely applied in the field of science, whose potential toxic effect has increasingly become a focus concern we need pay attention to in public health. The purpose of this article was to explore the toxicity mechanism with oxidative damage from treatment with QDs at the molecular level through a gene microarray.
Methods: Mice were administered aqueously synthesized cadmium telluride QDs (CdTe aqQDs) via intravenous tail injection of a 2 µmol/kg solution (based on the molar mass of Cd), and their kidneys were collected at 1 day in strict accordance with the programs used for treated mice. We determined the hierarchical clustering of expression ratios, enriched gene ontology (GO) terms and signaling pathways through gene microarray analysis and bioinformatics analysis in kidney tissue and screened the key enzyme genes, which were verified by real-time quantitative polymerase chain reaction (real-time qPCR).
Results: Compared to control group, 459 lncRNAs (197 down-regulated and 262 up-regulated) and 256 mRNAs (103 down-regulated and 153 up-regulated) were differentially expressed. According to biological processes in enriched GO terms, the response to a redox state played a significant role in the biological processes involved altered genes. Pathway analysis showed that the signaling pathways that involved cytochrome P450 (CYP450) enzymes had a close relationship with QDs. Among these signaling pathways, gene expression profiling revealed that selected differentially expressed mRNAs (CYP19A1, CYP1B1, CYP11A1, CYP11B2, and CYP17A1 in the kidney and CYP19A1 and CYP1B1 in the liver) were validated by real-time qPCR, resulting in expression levels of CYP11A1, CYP11B2 and CYP17A1 in the kidney and CYP19A1 and CYP1B1 in the liver that were significantly increased, however in expression levels of CYP19A1 and CYP1B1 compared with control group in the kidney, there was no significant difference.
Conclusions: Our results provide a foundation for and potential insight into the role of CYP450-related genes in QD-induced oxidative stress. QDs may produce a great deal of reactive oxygen species (ROS) by promoting high expression of CYP450 enzymes and accumulating steroid hormones, which may be an important toxicity mechanism for mediating oxidative stress and tissue damage.
Keywords: Quantum dots, Cytochrome P450 enzymes, Gene microarray, Oxidative stress
Introduction
Quantum dots (QDs) are one of semiconductor nanoparticles ranging from 2 to 20 nm in dimension, which are widely used in many fields, including molecular biology, cytobiology, biochemistry, and pharmacology, based on their characteristics of wide spectral range, high quantum yield, light stability, sensitivity and specificity [1-3]. Two strategies are used in synthesizing QDs: aqueous synthesis (aqQDs) and organic synthesis (orQDs). Compared with orQDs, aqQDs do not have large amounts of hydrophilic ligands to cover their surfaces and they are naturally water-dispersed. Moreover, aqQDs possess a much smaller hydrodynamic diameter than orQDs. Specifically, aqQDs are effective reagents because of good biocompatibility, which are compatible with biological system [4].
Cadmium telluride (CdTe) aqQDs are thought one of the most successful aqQDs, which have widespread use in fluorescence probes, drug targets and animal vivo imaging technology [5-7]. However, as is known to induce toxicity in organisms, cadmium (Cd) is the most abundant component of CdTe aqQDs. Therefore, numerous studies investigating the toxicity of CdTe aqQDs have been developed. Several published reports indicate that CdTe aqQDs could induce a reduction in the viability and motility of sperm in male mice [8]; exert genetic toxicity, such as gene mutation, chromosomal fragmentation and alteration on Hydra vulgaris [9]; mediate cell death through cell wall breakage and cytoplasm blebbing to exhibit marked cytotoxicity [10]; and cause an enlargement of mitochondria, a destruction of mitochondrial membrane potential, increasing intracellular calcium levels in HepG2 cells, etc. [11]. The toxicity of QDs may have multiple mechanisms, including the release of free metal ions at the core and the production of reactive oxygen species (ROS)-mediated oxidative stress [12-15].
ROS are a class of compounds, which have short half-lives and high reactivities based on their tendency to receive and give electrons to keep stability. They can be produced by the oxidation of a series of metabolic processes in cells, such as NAD (P) H oxidase system, xanthine oxidase system, arachidonic acid metabolism, monoamine oxidase system, mitochondrial respiratory chain and neutrophil respiratory burst of neutrophils and phagocytes, and the metabolism of extraneous substances by cytochrome P450 (CYP450) enzymes. Membrane lipids, proteins and DNA are the most vulnerable biological targets for ROS. Nevertheless, the overproduction of ROS can cause oxidative stress, even lead to irreversible damage in organelles, cells and tissues, although they play critical roles in metabolism. To maintain a production and consumption balance with ROS, the organisms could activate endogenous antioxidant systems, which contain superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GPx), glutathione (GSH), uric acid and ubiquinone, by ingestion of exogenous antioxidants and input from cofactors, with avoiding an excess of ROS [16-19].
The production of ROS was typically deduced on the basis of changes in antioxidant systems including lipid peroxidation levels and antioxidant enzyme activities, in which nanoparticles mediated oxidative stress in previous studies. Our previous study demonstrated that CdTe aqQDs appeared to be mainly deposited in kidney and liver tissues via tail intravenous administration in mice, CdTe aqQDs induced histopathological damages of the kidney and liver by elevating Cd2+, hydroxyl radicals (·OH), hepatocyte mitochondrial swelling, vacuole and crest reduction with time-dependent [20]. In order to understand the toxicity mechanism of CdTe aqQDs, we used the electron paramagnetic resonance (EPR) spin-trapping technique, which investigated the antioxidant capacities in the kidneys and livers of mice, and examined the activities of oxidative stress markers, which were SOD, CAT, GPx, GSH and MDA. Our research proved that CdTe aqQDs could deplete GSH to reduce the tissue total antioxidant capacity with the activities of SOD, CAT, GPx increasing [21]. However, in contrast with the sufficient studies regarding to antioxidant system induced by QDs, only a few oxidization system reports of QDs have been published.
Toxicogenomics, including gene microarrays and bioinformatics, has demonstrated its application for assessing toxicity and the mechanism of exogenous substances [22-24]. In the present study, kidneys of male ICR mice exposed to CdTe aqQDs were collected to screen the differentially expressed genes and signaling pathways using gene microarrays. Furthermore, the key genes were verified by real-time quantitative polymerase chain reaction (real-time qPCR) to explore the toxicity mechanism of oxidative damage treated with QDs at the molecular level.
Materials and methods
Characteristics of CdTe aqQDs
Thioglycolic acid-stabilized CdTe QDs were prepared by Nanjing University. Prior to use in our experiments, the CdTe QDs stock solutions were centrifuged at 500× g for 25 min at 20 ℃ prior to use. We dialyzed 3 h against 0.1% thioglycolate (sodium salt; Sigma-Aldrich, MO, USA) with above supernatants using a 10 kDa cellulose membrane (Sigma-Aldrich, MO, USA), and then continued to dialyze 2 h against distilled water at pH 8.3, what removed free Te, Cd and unbound thioglycolate [20]. We used inductively coupled plasma mass spectrometry (ICP-MS, 7500ce; Agilent Technologies, CA, USA) to quantitatively measure concentrations of Cd in CdTe aqQDs. We used atomic force microscope (AFM, Nano Scope 3D; Veeco, NY, USA) to analyse the CdTe aqQDs for surface characteristics and size distribution. What is more, we used fluorescence spectrometer (RF-5301; Shimadzu, Kyoto, Japan) to measure the CdTe aqQDs, which included peak wavelengths, fluorescence intensities and fluorescence spectra. The zeta-potential and hydrodynamic sizes of CdTe aqQDs were measured with a malvern instruments zetasizer (Nano-ZS90; Malvern Panalytical, Malvern, UK). The CdTe aqQD solutions were freshly dissolved in physiological saline with PBS (pH 7.4) and then sonicated within 10 min prior to injection into mice.
Animals
Male ICR mice, which were 6-8 weeks of age, provided from Beijing (Vital River Laboratory Animal Technology Co., Ltd., Beijing, China). All procedures and experimentations were approved by the Ethics Committee of Capital Medical University (ethical review number: AEEI-2016-184). These mice were raised in a standardized animal house at China Capital Medical University with a 12-hour day/dark revolution, in which operation was ventilated, sterile and temperature-controlled. Regular observation was conducted for each mouse containing fur, urine, mental status, behaviour, food intake and faces.
After 7 days of acclimatization, mice were administered the CdTe aqQD solutions by tail intravenous injection of 2 µmol/kg (based on the molar mass of Cd) between 33.1 and 35.8 g in weight, the control was injected physiological saline with an equivalent volume. Five mice from each exposed group were anaesthetized with isoflurane at the predetermined time points (1, 7 days), which were killed using cervical dislocation, their kidneys (1 day) were collected for gene microarray analysis, and the kidney and liver tissues (1, 7 days) were analysed with real-time qPCR. Sets of control mice were also killed at the predetermined times (1, 7 days) in strict accordance with the programs used for treated mice.
Gene microarray analysis
The tissue was cut into small pieces in vitro and quickly frozen in liquid nitrogen. The tissue sample was taken out of liquid nitrogen and the tissue block was placed in a pre-cooled mortar and ground. After the tissue sample was ground into a powder, 2-3 mL of TRIzol reagent (Invitrogen, Carlsbad, Canada) was added to each mortar and grinding was continued. The TRIzol reagent returned to the liquid state was transferred to a glass homogenizer. Homogenization was continued for 3-5 minutes. The TRIzol reagent was then transferred to a 1.5 mL centrifuge tube and the total RNA was subsequently extracted according to the Trizol reagent standard procedure. We used TRIzol reagent to extract the total RNA including lncRNA and mRNA, which were purified with a mirVana miRNA Isolation Kit (Ambion, TX, USA) following the instructions of manufacturer, from the kidneys (100 mg tissue) in mice. The quality and quantity of the RNA samples were measured from OD260/280 data with spectrophotometer (NanoDrop 2000; Thermo, MA, USA).
We used the GeneSpring software V13.0 (Agilent Technologies, CA, USA) to analyse the array data of lncRNA and mRNA about data normalization, quality control and summarization. Moreover, we utilized threshold values of ≥2 or ≤-2-fold change and p value of 0.05 in T-test as the standard for differentially expressed genes. The data were log2 transformed, median centered by genes using the Adjust Data function of CLUSTER 3.0 software and then further analysed with hierarchical clustering with average linkage [25].
Bioinformatics analysis
Gene ontology (GO) is a project that describes the function of genes and gene products in a unified definition. GO developed the standard language (ontologies) with tertiary structure. The biological process, cellular component and molecular functions were analysed simultaneously in enriched GO terms.
On the basis of Reactome database, pathway analysis, in which Fisher's exact and EASE-score tests were used, was applied to pick out the significant pathways by differentially expressed genes [26]. The threshold of significant pathways were analysed with FDR < 0.05.
Real-time qPCR analysis
Using a PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio Inc., Shiga, Japan), the cDNA was synthesized from 1.0 μg of total RNA. The mRNA levels of CYP450-related genes were determined by real-time qPCR (Mastercycler; Eppendorf, Hamburg, Germany) according to the protocols of manufacturer. Primers (the sequences of the forward and reverse primers used in this experiment are summarized in Table 1) for the CYP450 enzymes and reference gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were designed by the Sangon Company. The comparative CT method was used for quantification, with GAPDH as a housekeeping gene, and the relative abundances were normalized to the control.
Statistical analysis
Statistics analysis were performed by One-way analysis of variance (ANOVA) using SPSS 19.0 software, followed by the least significant difference (LSD) test. Data were expressed as the mean±SD, and FDR < 0.05 was set for significance level.
Primers used for Real-time qPCR.
| Isoenzymes | Forward | Reverse |
|---|---|---|
| GAPDH | CGTGTTCCTACCCCCAATG | ATGTCATCATACTTGGCAGGTT |
| CYP1B1 | CACCAGCCTTAGTGCAGACAG | GAGGACCACGGTTTCCGTTG |
| CYP19A1 | ATGTTCTTGGAAATGCTGAACCC | AGGACCTGGTATTGAAGACGAG |
| CYP17A1 | AGTTTGCCATCCCGAAGGA | CTGGCTGGTCCCATTCATTT |
| CYP11A1 | GGCCCAGCGGTTCATCAAT | TGCCTTAAGTCCCAGTAGA |
| CYP11B2 | GCCACCATGGCACTCAGGGCAAAGGCAGAGG | CTAGTTAATCGCTCTGAAAGTGAGGAGGGGGGACG |
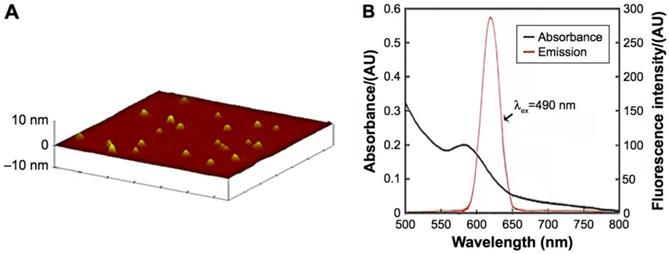
Characteristics of the CdTe aqQDs. Notes: (A) AFM image; (B) absorption and emission spectra. The diameter of the CdTe aqQDs was 3-4 nm. The excitation occurred at 490 nm, the maximum emission occurred at 620 nm [20, 21].
Results
Characteristics of CdTe aqQDs
ICP-MS indicated that the concentration of the CdTe aqQDs was 5 µmol/mL (based on the molar mass of Cd). AFM, which showed that the diameter of the CdTe aqQDs was approximately 3-4 nm, was used to evaluation in size. Fluorescence spectrum suggested that according to excitation at 490 nm, the maximum emission occurred at 620 nm (Fig. 1). This part is the preliminary work of our research group [21]. The zeta-potential value of the CdTe aqQDs dispersed in water is (-42.6±4.8) mV. Hydrodynamic size was performed by dynamic light scattering (DLS), the sizes of the CdTe aqQDs were determined to be (132.4±24.8) nm.
Differential gene expression induced by CdTe aqQDs
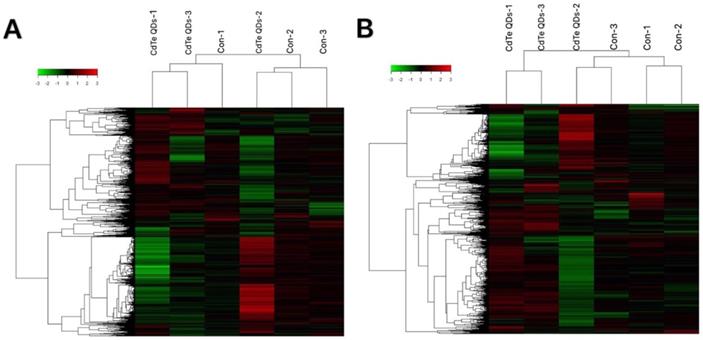
To understand which sensitive RNAs induced by CdTe aqQDs are involved in the kidney, gene microarray analysis was performed in the CdTe aqQDs group and control group (three samples per group). We assessed the expression profiles of lncRNA and mRNA with an lncRNA and mRNA mice gene expression microarray. Compared to control group, a total of 459 lncRNAs were differentially expressed in the CdTe aqQDs-induced kidney group, in which 197 lncRNAs were down-regulated and 262 lncRNAs were up-regulated. Additionally, a total of 256 mRNAs were differentially expressed in the CdTe aqQDs-induced kidney group: 103 mRNAs were down-regulated, 153 mRNAs were up-regulated (Fig. 2).
GO analysis induced by CdTe aqQDs
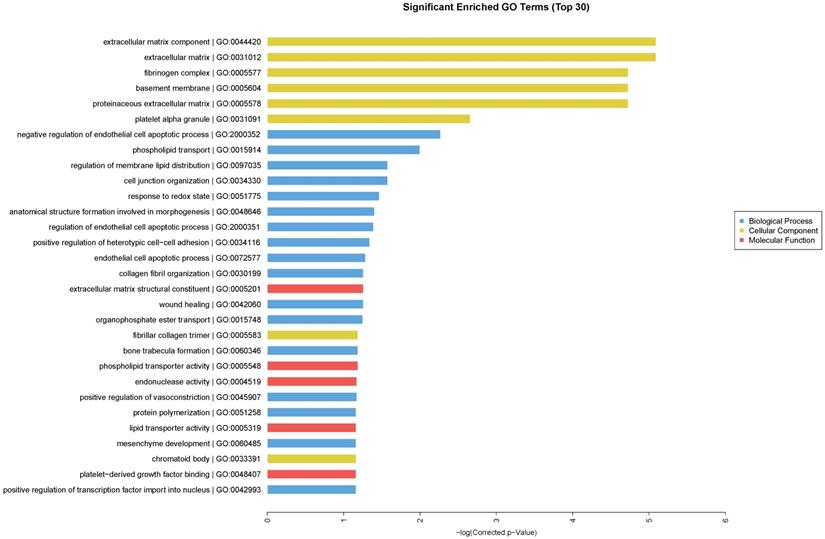
We performed GO enrichment analysis, which are arranged and displayed according to biological processes, molecular functions and cellular components (Fig. 3), to identify the gene expression changes associated with CdTe aqQDs treatment. The significantly enriched categories of our gene set were extracellular component, negative regulation of endothelial cell apoptotic process, extracellular matrix structural constituent, and regulation of membrane lipid distribution. Especially among these, we were interested to find that the response to redox state played a significant role in the biological processes involved in these altered genes. The biological process of redox regulation serves to protect the body from oxidative damage, maintaining a dynamic balance.
Pathway analysis induced by CdTe aqQDs
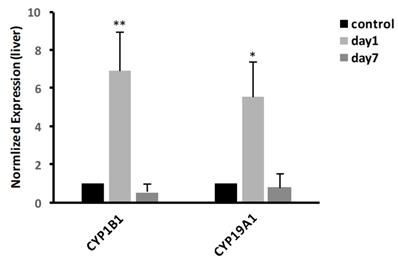
We determined the biological pathways related to the most differentially expressed RNAs in kidney using pathway analysis with the Reactome database. The method calculated the significance of gene enrichment in each pathway entry and further classified the signaling pathways involved in the genetic variations. The results, as shown in Fig. 4, indicated that CdTe aqQDs could cause change in mice with 65 signaling pathways involved in the sensitive-expressed RNAs. Among these, Defective CYP26C1 causes focal facial dermal dysplasia 4, Defective CYP1B1 causes glaucoma, Defective CYP17A1 causes adrenal hyperplasia 5, Defective CYP11A1 causes congenital adrenal insufficiency, with 46, XY sex reversal, mainly related to CYP450-related genes. Therefore, we found that CYP450-related signaling pathways were sensitive-induced by CdTe aqQDs.
RNA expression profiles between CdTe aqQDs-induced group and normal group in kidney tissues. Notes: Hierarchical clustering of (A) lncRNAs; (B) mRNAs with expression ratios (log2 scale), “green” represents a low relative expression and “Red” represents a high relative expression.
Enriched GO terms according to biological processes, molecular functions and cellular components. Notes: “blue” represents biological process; “yellow” represents cellular component; “red” represents molecular function. Differentially expressed transcripts were included with P<0.05 and fold-change>2.
Effects of CdTe aqQDs on mRNA expression related to CYP450 enzymes in kidney
To validate the microarray results, we selected the CYP450-related genes in some of the significantly enriched signaling pathways. The results in kidney, as shown in Fig. 5, demonstrated that compared with control group, the expression of CYP17A1 was significantly increased to 5.37-fold (P<0.01) at 1 day. Moreover, compared with control group, the expression of CYP11A1 was significantly increased to 6.61-fold (P<0.05), while the expression of CYP11B2 was significantly increased to 3.32-fold (P<0.05). At 7 days, the gene expression levels of CYP17A1, CYP11A1 and CYP11B2 were decreased to the same level as that in control group. Compared with control group, no significant differences were found in expression of CYP19A1 and CYP1B1 (P>0.05) at 1 and 7 days. In general, compared to control, the mRNA expression levels of CYP17A1, CYP11A1 and CYP11B2 were increased. However, the mRNA expression levels of CYP19A1 and CYP1B1 were unchanged.
Effects of CdTe aqQDs on mRNA expression related to CYP450 enzymes in liver
In liver, we examined the mRNA expressions of CYP19A1 and CYP1B1. The result, as shown in Fig. 6, showed that at 1 day, the expression of CYP19A1 was significantly increased 5.55-fold (P<0.05); meanwhile, compared with control group, the expression of CYP1B1 was significantly increased to 6.9-fold (P<0.01). At 7 days, the gene expression levels of CYP19A1 and CYP1B1 were decreased to the same level as that in control group.
Signaling pathways of differentially expressed RNAs. Notes: The horizontal axis represents the -log 10 (p value) of these significant pathways and the vertical axis represents the pathway category. The two-sided Fisher's exact test with P<0.05 was significant in classification.
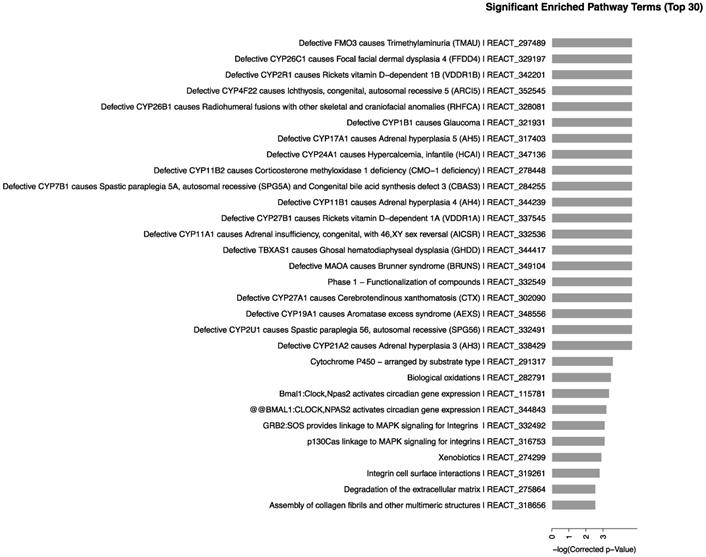
Effects of CdTe aqQDs on the mRNA expressions of CYP1B1, CYP19A1, CYP11A1, CYP11B2 and CYP17A1 in the kidney at 1 and 7th day. Notes: *P<0.05 vs. control; **P<0.01 vs control.
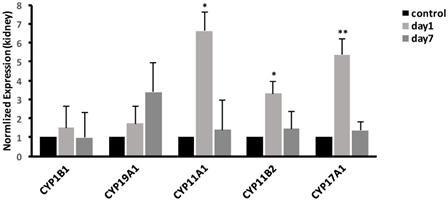
Effects of CdTe aqQDs on the mRNA expressions of CYP1B1 and CYP19A1 in the liver at 1 and 7th day. Notes: *P<0.05 vs. control; **P<0.01 vs control.
Discussion
With the wide application of QDs in the field of life science, their potential toxic effect has increasingly become one of the focuses of public health concern. Researches have showed that QDs distribute in multiple organs and are mainly accumulated in the liver and kidney, resulting in damage to those organs [27-29]. As a kind of advanced, large-scale and high-throughput detection technology, gene microarrays have been widely used in many fields, such as disease diagnosis and treatment, drug screening, and environmental testing [30-32]. To further study the molecular mechanism of damage induced by CdTe aqQDs, male ICR mice were selected as the experimental subjects; kidneys of mice were collected to screen the differentially expressed genes and signaling pathways using gene microarray. The results showed that the expression of 459 lncRNA genes changed after CdTe aqQDs exposure, including 197 down-regulated genes and 262 up-regulated genes; the expression of 256 mRNA genes changed, including 103 down-regulated genes and 153 up-regulated genes.
GO analysis is the classical method to organize differentially expressed genes into hierarchical categories based on gene function [33]. According to CdTe aqQDs-induced function enriched classification, the response to redox state played an important role in the biological processes involved in genes that were differentially altered. Subsequently, we performed pathway analysis in differentially expressed genes based on the Reactome database, and the results showed that CdTe aqQDs caused the changes of 65 signaling pathways in the kidney, among which the markedly enriched signaling pathways are mainly CYP450-related signaling pathways. CYP450 enzymes is widely distributed in animals, plants and microorganisms, as a kind of multifunctional enzyme, which mainly exists in liver microsome and is a large superfamily of genes composed of many enzymes. These enzymes participate in the phase I reaction of endogenous and exogenous compounds in liver degradation and play an important role in biotransformation in vivo [34]. CYP450 enzymes, a major protein binding to the mitochondrial membrane, is responsible for the transfer of electrons throughout the metabolic process and is an important site for the production of ROS [35,36].
Studies both at home and abroad show that the toxicity mechanism mainly focuses on the release of free Cd2+ and the generation of ROS, which mediate the oxidative stress [3,14,15,37]. In recent years, our group found that CdTe aqQDs could degrade and release Cd2+ after they entered mice [20]. Studies in the literature showed that Cd2+ increased the expression of CYP450 enzymes and intracellular ROS concentration triggered oxidative stress in the body and caused damages to the liver and kidney [38,39]. Therefore, the damage by CdTe aqQDs may be caused by the degradation of Cd2+ after CdTe aqQDs entered the body, resulting in the accumulation of ROS. According to the above, combined with the results of the gene microarray, this study suggests that the CdTe aqQDs-induced injury in the liver and kidney may be caused by Cd2+ degraded from CdTe aqQDs, the high expression of CYP450 enzymes activity, and then the triggering of a large number of ROS, which may be an important mechanism mediating oxidative stress and tissue injury.
To further verify the above results, we selected the CYP450-related genes in some of the significantly enriched signaling pathways and detected the expression of these key genes in the liver and kidney using real-time qPCR. The results showed that the expression levels of CYP17A1, CYP11A1 and CYP11B2 in the kidney were significantly increased; expression levels of CYP19A1 and CYP1B1 in the liver were significantly increased, but compared with control group, there was no significant difference in the kidney. Studies in the literature studies showed that these differentially expressed genes are key genes for metabolism of steroid hormones. CYP11A1 is the first rate-limiting enzyme in the biosynthetic pathway of steroid hormone, which converts cholesterol into pregnenolone and is a precursor of progesterone biosynthesis [40]. CYP17A1 converts progesterone to androstenedione, which is a direct precursor of testosterone biosynthesis [40]. CYP19A1 converts testosterone to oestradiol and hydroxylation of oestradiol by CYP1B1 to produce 4-OH-oestradiol and 2-OH-oestradiol [41]. CYP1B1 can further convert 4-OH-oestradiol and 2-OH-oestradiol into quinone and semiquinone, which can produce ROS through redox cycle, resulting in oxidative damage [42]. Additionally, the research of Fedotcheva T on steroid hormones also revealed that steroid hormones regulated the production of ROS [43]. Therefore, it is speculated that CdTe aqQDs may cause the accumulation of steroid hormones through up-regulation of the genes involved in steroid synthesis and produce a large amount of ROS, which causes the damages to the liver and kidney in the body.
Conclusions
In summary, the results of this study showed that CdTe aqQDs induced differential expressions of mRNA and lncRNA in the liver and kidney of mice. CYP450-related signaling pathways are key pathways leading to the damages of liver and kidney tissues. The high expression of CYP450 enzymes and the accumulation of steroid hormones led to the generation of a large number of ROS, which was an important mechanism of CdTe aqQDs-mediated oxidative stress and tissue damage. Meanwhile, our research also provided the experimental evidence for further elucidating the toxic effects and molecular mechanisms of CdTe aqQDs on biological metabolic systems in the future.
Acknowledgements
We thank Han Ying and Lihong Zhou for helpful discussions. This work was supported by the National Natural Science Foundation of China (81573201, 81273131).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bruchez MJ, Moronne M, Gin P. et al. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013-6
2. Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016-8
3. Derfus AM, Chan WCW, Bhatia SN. Probing the cytotoxicity of semiconductor quantum dots. Nano Lett. 2004;4:11-18
4. Liu N, Mu Y, Chen Y. et al. Degradation of aqueous synthesized CdTe/ZnS quantum dots in mice: differential blood kinetics and biodistribution of cadmium and tellurium. Part Fibre Toxicol. 2013;10:37
5. Larson DR, Zipfel WR, Williams RM. et al. Water-soluble quantum dots for multiphoton fluorescence imaging in vivo. Science. 2003;300:1434-6
6. Guszpit E, Krejcova L, Krizkova S. et al. Kinetic analysis of human metallothionein and CdTe quantum dot complexes using fluorescence and voltammetry techniques. Colloids Surf B Biointerfaces. 2017;160:381-9
7. Xiao J, Bai Y, Wang Y. et al. Systematic investigation of the influence of CdTe QDs size on the toxic interaction with human serum albumin by fluorescence quenching method. Spectrochim Acta A Mol Biomol Spectrosc. 2010;76:93-97
8. Akhavan O, Hashemi E, Zare H. et al. Influence of heavy nanocrystals on spermatozoa and fertility of mammals. Mater Sci Eng C Mater Biol Appl. 2016;69:52-59
9. Ambrosone Alfredo, Mattera L et al. Mechanisms underlying toxicity induced by CdTe quantum dots determined in an invertebrate model organism. Biomaterials. 2012;33:1991-2000
10. Han X, Lai L, Tian F. et al. Toxicity of CdTe quantum dots on yeast Saccharomyces cerevisiae. Small. 2012;8:2680-9
11. Nguyen KC, Rippstein P, Tayabali AF. et al. Mitochondrial toxicity of cadmium telluride quantum dot nanoparticles in mammalian hepatocytes. Toxicol Sci. 2015;146:31-42
12. Kirchner C, Javier AM, Susha AS. et al. Cytotoxicity of nanoparticle-loaded polymer capsules. Talanta. 2005;67:486-91
13. Su Y, Hu M, Fan C. et al. The cytotoxicity of CdTe quantum dots and the relative contributions from released cadmium ions and nanoparticle properties. Biomaterials. 2010;31:4829-34
14. Tang S, Cai Q, Chibli H. et al. Cadmium sulfate and CdTe-quantum dots alter DNA repair in zebrafish (Danio rerio) liver cells. Toxicol Appl Pharmacol. 2013;272:443-52
15. Katsumiti A, Gilliland D, Arostegui I. et al. Cytotoxicity and cellular mechanisms involved in the toxicity of CdS quantum dots in hemocytes and gill cells of the mussel Mytilus galloprovincialis. Aquat Toxicol. 2014;153:39-52
16. Halliwell B. Free radicals and antioxidants - quo vadis?. Trends Pharmacol Sci. 2011;32:125-30
17. Wu JQ, Kosten TR, Zhang XY. Free radicals, antioxidant defense systems, and schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2013;46:200-6
18. Rahal A, Kumar A, Singh V. et al. Oxidative stress, prooxidants, and antioxidants: the interplay. Biomed Res Int. 2014;2014:761264
19. Niki E. Assessment of antioxidant capacity in vitro and in vivo. Free Radic Biol Med. 2010;49:503-15
20. Wang M, Wang J, Sun H. et al. Time-dependent toxicity of cadmium telluride quantum dots on liver and kidneys in mice: histopathological changes with elevated free cadmium ions and hydroxyl radicals. Int J Nanomedicine. 2016;11:2319-28
21. Wang J, Sun H, Meng P. et al. Dose and time effect of CdTe quantum dots on antioxidant capacities of the liver and kidneys in mice. Int J Nanomedicine. 2017;12:6425-35
22. Robinson JF, Griffith WC, Yu X. et al. Methylmercury induced toxicogenomic response in C57 and SWV mouse embryos undergoing neural tube closure. Reprod Toxicol. 2010;30:284-91
23. Robinson JF, Yu X, Moreira EG. et al. Arsenic- and cadmium-induced toxicogenomic response in mouse embryos undergoing neurulation. Toxicol Appl Pharmacol. 2011;250:117-29
24. Kwon JY, Weon JI, Koedrith. et al. Identification of molecular candidates and interaction networks via integrative toxicogenomic analysis in a human cell line following low-dose exposure to the carcinogenic metals cadmium and nickel. Oncol Rep. 2013;30:1185-94
25. Eisen MB, Spellman PT, Brown PO. et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863-8
26. Hosack DA, Dennis GJ, Sherman BT. et al. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70
27. Han Y, Xie G, Sun Z. et al. Plasma kinetics and biodistribution of water-soluble CdTe quantum dots in mice: a comparison between Cd and Te. J Nanopart Res. 2011;13:5373-80
28. Ott M, Gogvadze V, Orrenius S. et al. Mitochondria, oxidative stress and cell death. Apoptosis. 2007;12:913-22
29. Szuster-Ciesielska A, Stachura A, Slotwinska M. et al. The inhibitory effect of zinc on cadmium-induced cell apoptosis and reactive oxygen species (ROS) production in cell cultures. Toxicology. 2000;145:159-71
30. Lupien M, Eacute J, Ocirc R. et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription: Cell. Cell. 2008;132:958-70
31. T.D. Harris, P.R. Buzby, H. Babcock, E. Beer, J. Bowers, I. Braslavsky, et al, Single-molecule DNA sequencing of a viral genome, Science. 2008;320:106-9
32. Liu JH, Chen G, Dang YW. et al. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev. 2014;15:2971-7
33. Lu C, Xiong M, Luo Y. et al. Genome-wide transcriptional analysis of apoptosis-related genes and pathways regulated by H2AX in lung cancer A549 cells. Apoptosis. 2013;18:1039-47
34. Romoser AA, Chen PL, Berg JM. et al. Quantum dots trigger immunomodulation of the NFkappaB pathway in human skin cells. Mol Immunol. 2011;48:1349-59
35. Nieto N, Friedman SL, Cederbaum AI. Stimulation and proliferation of primary rat hepatic stellate cells by cytochrome P450 2E1-derived reactive oxygen species. Hepatology. 2002;35:62-73
36. Sakurai K, Cederbaum AI. Oxidative stress and cytotoxicity induced by ferric-nitrilotriacetate in HepG2 cells that express cytochrome P450 2E1. Mol Pharmacol. 1998;54:1024-35
37. Zhao Y, Lin K, Zhang W. et al. Quantum dots enhance Cu2+ -induced hepatic L02 cells toxicity. J Environ Sci. 2010;22:1987-92
38. Satarug S, Nishijo M, Lasker JM. et al. Kidney dysfunction and hypertension: role for cadmium, p450 and heme oxygenases?. Tohoku J Exp Med. 2006;208:179-202
39. Sinha M, Manna P, Sil PC. Induction of necrosis in cadmium-induced hepatic oxidative stress and its prevention by the prophylactic properties of taurine. J Trace Elem Med Biol. 2009;23:300-13
40. Toda K, Hayashi Y, Ono M. et al. Co-administration of insulin with a gonadotropin partly improves ovulatory responses of estrogen-deficient mice. Mol Cell Endocrinol. 2015;411:177-86
41. Lee SR, Lee SY, Kim SY. et al. Hydroxylation and sulfation of sex steroid hormones in inflammatory liver. J Biomed Res. 2017;31:437-44
42. Li F, Zhu W, Gonzalez FJ. Potential role of CYP1B1 in the development and treatment of metabolic diseases. Pharmacol Ther. 2017;178:18-30
43. Fedotcheva TA, Kruglov AG, Teplova VV. et al. [Effect of steroid hormones on production of reactive oxygen species in mitochondria]. Biofizika. 2012;57:1014-9
Author contact
![]() Corresponding author: Prof. Peili Huang, School of Public Health, Capital Medical University, No 10 Xitoutiao, You An Men, Beijing 100069, China. Tel: +86 10 8391 6539; Fax: +86 10 8391 1507; Email: huangpledu.cn
Corresponding author: Prof. Peili Huang, School of Public Health, Capital Medical University, No 10 Xitoutiao, You An Men, Beijing 100069, China. Tel: +86 10 8391 6539; Fax: +86 10 8391 1507; Email: huangpledu.cn
Received 2017-12-27
Accepted 2018-6-7
Published 2018-6-19
