ISSN: 2206-7418
Nanotheranostics 2017; 1(1):80-102. doi:10.7150/ntno.18216 This issue Cite
Review
Gold Nanoparticle Based Platforms for Circulating Cancer Marker Detection
Department of Chemistry, The University of Memphis, Memphis, TN 38152, USA.
Received 2016-11-5; Accepted 2016-12-22; Published 2017-1-19
Abstract
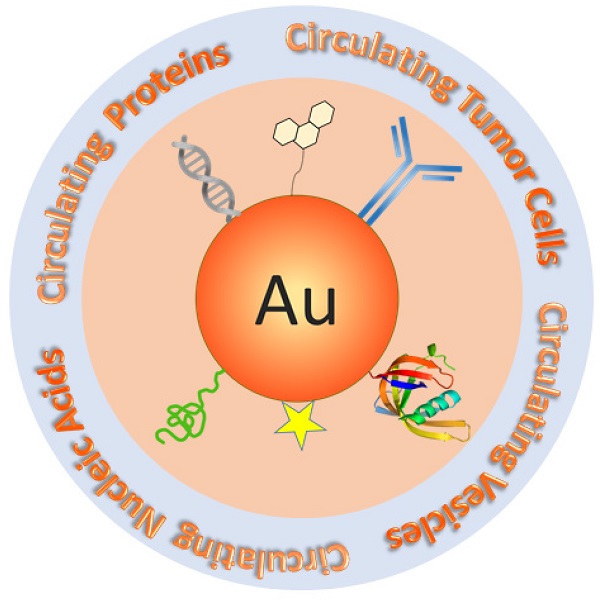
Detection of cancer-related circulating biomarkers in body fluids has become a cutting-edge technology that has the potential to noninvasively screen cancer, diagnose cancer at early stage, monitor tumor progression, and evaluate therapy responses. Traditional molecular and cellular detection methods are either insensitive for early cancer intervention or technically costly and complicated making them impractical for typical clinical settings. Due to their exceptional structural and functional properties that are not available from bulk materials or discrete molecules, nanotechnology is opening new horizons for low cost, rapid, highly sensitive, and highly specific detection of circulating cancer markers. Gold nanoparticles have emerged as a unique nanoplatform for circulating biomarker detection owning to their advantages of easy synthesis, facile surface chemistry, excellent biocompatibility, and remarkable structure and environment sensitive optical properties. In this review, we introduce current gold nanoparticle-based technology platforms for the detection of four major classes of circulating cancer markers - circulating tumor cells, vesicles, nucleic acids, and proteins. The techniques will be summarized in terms of signal detection strategies. Distinctive examples are provided to highlight the state-of-the-art technologies that significantly advance basic and clinical cancer research.
Keywords: Gold nanoparticle, circulating cancer marker detection, circulating tumor cell, extracellular vesicle, circulating nucleic acid, protein tumor marker.
Introduction
Tissue biopsy is commonly used in the clinic to diagnose a variety of cancers. It plays an important role in tailoring best treatment options for the individuals at diagnosis. However, tissue-based biopsy is invasive, quite costly, impractical for repeated testing, and unavailable to some cancer types. For example, over 30% of advanced non-small cell lung cancers do not have accessible tissue [1]. Especially, tumors are heterogeneous and evolve over time. Thus, biopsy data from the limited amount of collected tissues are often biased and can mislead clinical decisions.
The limitations in tissue biopsy have driven the development of liquid biopsy, a means to detect and analyze cancer biomarkers in body fluids such as blood and urine to aid in cancer screening, diagnosis and therapy [2]. Liquid biopsy is non-invasive, inexpensive, accessible to large populations, and allows for repeated testing for real-time monitoring of disease stage and treatment response. It is probably better than standard biopsies to assess tumor biology as biofluid samples contain information from all the tumor cells in the patient rather than small portion of a tumor in the tissue biopsy. It can detect minimal tumor residues and recurrence after surgery and has the potential to detect cancer before tumor identification with imaging techniques [3]. In June 2016, the U. S. Food and Drug Administration (FDA) approved the first liquid biopsy test, cobas epidermal growth factor receptor (EGFR) Mutation Test v2 (Roche Molecular Systems, Inc.) for in vitro diagnostic use in cancer. This test uses plasma specimens as a companion diagnostic test to detect defined mutations of the EGFR gene that makes patients with non-small cell lung cancer for treatment with the targeted therapy erlotinib. Liquid biopsies via comprehensive molecular profiling are already available to physicians and patients, which give a great source of additional tumor genetic information to tissue biopsy.
There are four classes of analytes in liquid biopsy: circulating tumor cells (CTCs), circulating vesicles, circulating nucleic acids (CNAs), and circulating proteins. Circulating vesicles consist mainly of exosomes (EXOs) and microvesicles (MVs) and CNAs of circulating tumor DNA (ctDNA) and RNA. It is still unclear which of these analytes is the best cancer biomarker. Most likely, a combination is needed to assess various aspects of cancer. Detection and analysis of circulating cancer biomarkers, however, is challenging because they represent small fractions in the complicated body fluids. For example, as few as one CTC is mixed with ~ 7 million white blood cells (WBCs) and 5 billion red blood cells (RBCs) in 1 mL of patient blood [4]. The fraction of ctDNA is often less than 1% (sometimes less than 0.01%) of total cell free DNA in patient plasma [5, 6]. The circulating vesicles from tumor cells are abundant (108 - 109 /mL of plasma), but non-tumor cells also release vesicles as part of their normal functions. The plasma may contain as many as 40,000 different proteins from about 500 gene products, which places a grand challenge to look for one or few specific oncoproteins. Therefore, detection and analysis require highly sensitive and specific techniques to identify and detect circulating biomarkers with high efficiency. Circulating biomarkers can be detected either by protein-based or nucleic acid-based approaches. Traditional protein-based methods are western blot, enzyme-linked immunosorbent assay (ELISA), radioimmunoassay (RIA), flow cytometry, mass spectrometry, and immunofluorescence imaging. Nucleic acid-based methods are polymerase chain reaction (PCR) and reverse transcription polymerase chain reaction (RT-PCR). Although these methods have made great advancements for circulating biomarker detection, they either lack sufficient sensitivity for early detection or are technically complicated requiring expensive instrumentation and skilled professionals. For example, the blood concentrations of proteins associated with early stage cancers range from 10-16 to 10-12 M [7], but the commercially available immunoassays have a typical limit of detection (LOD) at the picomolar level [8] rendering them incapable of early detection. It is therefore very important to develop new quantitative assays with ultrasensitivity.
Nanotechnology is opening new horizons for highly sensitive and specific detection of circulating cancer markers. The rationale is that nanomaterials exhibit exceptional functional properties that are often not available from either bulk materials or discrete molecules. Nanomaterials have large surface-to-volume ratio for highly efficient target interactions. These properties can be exploited to enhance the performance of traditional methods or develop new assays with ultrasensitivity and multiparametric capabilities. Nanosensors have reached detection limit from picomolar to zeptomolar levels, which opens a new era of early cancer detection [9]. Among the various nanoplatforms, gold nanoparticles (Au NPs) are unique for biomarker detection due to their easy synthesis, facile surface chemistry, excellent biocompatibility, and especially their remarkable optical properties. Like other noble metal NPs, Au NPs exhibit strong localized surface plasmon resonance (LSPR), the collective oscillation of conduction electrons around the particles that are induced by the electric field of incident light [10]. The LSPR of Au NPs is size, shape, structure, inter-particle distance, and environment sensitive, with tunable wavelength from visible to near infrared (NIR) regions [11-13]. Due to the LSPR, Au NPs exhibit extremely high absorption and scattering properties, with extinction coefficients on the orders of 108 to 1011 M-1cm-1 depending on the particle's size, shape and structure [14, 15]. These values are more than 10,000 times stronger than those of organic dyes. The LSPR also strongly enhances the electric fields of the incident and scattering light, with |E|2 being 100 to 10,000 times greater in magnitude than the incident field [16]. A direct outcome of this field enhancement is the strong augmentation of the Raman signals of adsorbed molecules as the intensity of Raman signals is proportional to the fourth power of the local field of the metal particle [17]. Additionally, Au NPs can modulate the fluorescence properties of adjacent species via radiative and nonradiative processes [18]. These optical properties are well suited to develop ultrasensitive platforms for detection and analysis of circulating cancer biomarkers. For example, a nanoplasmonic sensor composed of periodic Au nanohole arrays can detect 670 aM exosomes. This sensitivity is 104 times higher than western blot and 102 higher than ELISA [19]. Au NP-based bio-barcode assay can detect protein markers with LOD 4 - 6 orders of magnitude lower than ELISA [20].
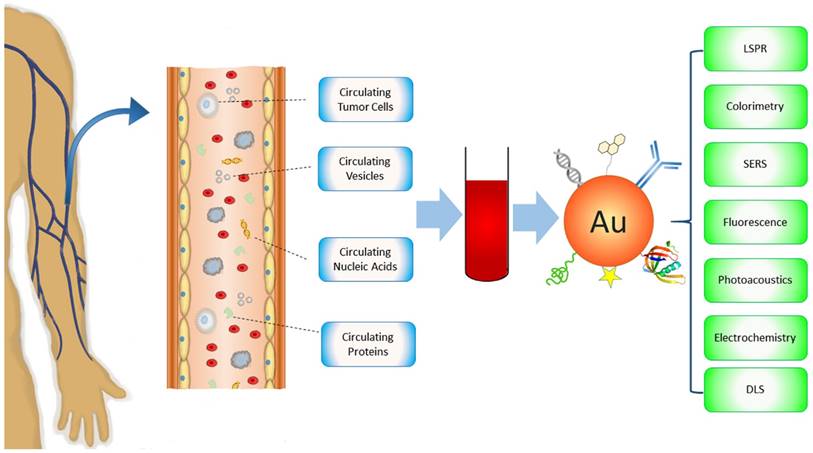
There are many reviews on Au NPs regarding their applications for cancer diagnosis and therapy [21-25]. However, none of them focus on circulating cancer markers, a rapidly growing field of cancer medicine. In this review, we introduce current Au NP-based technology platforms for the detection of circulating cancer markers categorized by the types of targets - CTCs, circulating vesicles, CNAs and circulating proteins (Figure 1). The techniques will be discussed in terms of signal detection mechanisms, including LSPR, colorimetric, surface enhanced Raman scattering (SERS), fluorescence, photoacoustic, electrochemical, and dynamic light scattering (DLS) detections. Other detection methods such as X-Ray imaging and MS will also be described. Distinctive examples, including those that have been tested with real patient samples (Table 1), are provided to highlight the state-of-the-art of Au NP-based technologies that significantly advance basic and clinical cancer research.
Schematic of Au NP-based platforms for the detection of circulating cancer markers including circulating tumor cells, circulating vesicles, circulating nucleic acids, and circulating proteins.
Summary of Au NP-based assays that have been tested with clinical samples for circulating cancer marker detection.
| Biomarkers | Nanomaterials | Target | Detection method | Detection limit | Linear range | Type of cancer | Ref |
|---|---|---|---|---|---|---|---|
| Circulating tumor cells | Au NPs | whole cell | SERS | 5 CTCs / mL blood | 10-1000 | head and neck | [57] |
| Au NPs | non-coding RNA | nanowire resonance frequency shift | 1 CTC / 10 mL blood | N/A | prostate | [87] | |
| Au NPs | DNA | fluorescence | 1 CTC / mL blood | 1-100 | colorectal | [88] | |
| Au NPs | mRNA | fluorescence | 100 CTCs / mL blood | 102 - 105 | breast | [92] | |
| Circulating vesicles | Au nanohole array | exosome | LSPR | 670 aM | 104 - 106 | ovarian | [19] |
| Au NPs | exosome | colorimetric | 1.42 pM | 1.4 pM-2.2 nM | melanoma | [133] | |
| Circulating nucleic acids | Au NPs | ctDNA | SERS | 10 copies | 10-10,000 copies | melanoma | [161] |
| Circulating proteins | Au NPs | CEA | colorimetric | 34 pg/mL | 0.05-20 ng/mL | colorectal | [185] |
| Au NPs | AFP | colorimetric | 2 ng/mL | 2-80 ng/mL | liver | [188] | |
| Au NPs | PSA | colorimetric | 4.6 fg/mL | 10-105 fg/mL | prostate | [191] | |
| Au NPs | PSA | bio-barcode | 330 fg/mL | 0.33-33 pg/mL | prostate | [195] | |
| Au NSTs | CA15.3, CA27.29 & CEA | SERS | 0.05 ng/mL | 0.01-103 ng/mL | breast | [214] | |
| Au NSTs | VEGF | SERS | 7 fg/mL | 0.1-104 pg/mL | breast | [215] | |
| Au NPs | MUC4 | SERS | N/A | N/A | prostate | [216] | |
| HGNs | CEA & AFP | SERS | 1 ng/mL | 1-100 ng/mL | liver | [218] | |
| Au NPs | MUC4 | SERS | 33 ng/mL | 0.01-10 ug/mL | pancreatic | [219] | |
| Au NPs | serum biochemicals | SERS | N/A | N/A | colorectal | [221] | |
| Au NPs | serum biochemicals | SERS | N/A | N/A | nasopharyngeal | [222] | |
| Au NPs | HER2 | electrochemical | 7.4 ng/mL | 10-110 ng/mL | breast | [226] | |
| Au NPs | CEA&AFP | electrochemical | 3 pg/mL | 0.01-60 ng/mL | liver | [229] | |
| Au NPs | EGFR | electrochemical | 50 pg/mL | 1-40 ng/mL | breast | [231] | |
| Au NPs | IgG | DLS | N/A | N/A | prostate | [234] |
Circulating Tumor Cells (CTCs)
CTCs are a hallmark of invasive behavior of cancer. Based on the “seed and soil” theory, a small portion of CTCs (<0.01%) arrest in a capillary bed at a distant site where they extravagate and seed the growth of a secondary tumor [26]. CTCs are therefore a required step for metastasis. Although the clinical values of CTCs remain unclear, many studies have shown their great potential [27]. CTCs can be used to characterize and monitor cancer progression [28]. The prognostic significance of CTCs has been demonstrated in several types of cancers including breast, prostate, colon, melanoma, and lung cancer [4, 29-32]. CTCs are also useful in monitoring and predicting the responses to ongoing therapy [33-35]. In addition, detection of CTCs shows strong promise for early cancer detection since CTCs have been found in blood during early stages of tumorigenesis [36]. CTCs are rare events, as few as one CTC mixed with 10 million WBCs and 5 billion RBCs in 1 mL of patient blood with advanced cancer [4]. Thus, it requires highly sensitive and specific techniques to capture and identify them with high efficiency. To date, a vast number of isolation and detection techniques have been developed, with about 100 companies offering CTC-related products and devices and over 400 ongoing clinical trials [37]. However, only one technique, the CellSearch system (Veridex, LLC), has been cleared by FDA for clinical utilization. This technique uses 120-200 nm iron (Fe) NPs (ferrofluid) linked with epithelial cell adhesion molecule (EpCAM) antibodies to magnetically isolate CTCs from blood plasma and then detect the tumor cells with immunofluorescence imaging based on intracellular cytokeratin expression [38]. EpCAM is an epithelia marker. Since normal epithelial cells are not found in blood circulation, capturing EpCAM-positive cells indicates the capture of CTCs. The CellSearch system is used to detect and count CTCs in blood of patients with breast, prostate, and colon cancers. However, this technique has poor sensitivity, typically detecting CTCs in only 50% patients known to have metastasis [39-41].
Au NPs can be used to capture CTCs by making two-dimension nanostructured substrates, either periodic Au nanostructure array [42] or Au NP-deposited surface [43-46]. Targeting ligands such as anti-EpCAM antibodies and aptamers are coated on the Au surface to bind and capture CTCs. Non-targeted blood cells and other impurities are easily washed away from the substrate with buffer solution. Due to the enhanced cellular interaction from the large surface-to-volume ratio, the nanostructured substrate can capture over 90 % CTCs in contrast to 49 % with solid surface [45]. Using the photothermal properties of NIR-absorbing Au nanorods (NRs) in conjunction with a thermo-responsive hydrogel substrate, the captured CTCs can be released at single cell resolution for downstream analysis [46]. A very recent approach is to coat anodic and cathodic electrodes with Au NPs to capture CTCs for subsequent electrochemiluminesent detection [47]. By grafting the cathodic Au NPs with EpCAM aptamer and anodic Au NPs with lectin, this approach can capture CTCs and profile surface glycan expressions by utilizing the changes of the anodic to cathodic peak intensity ratio.
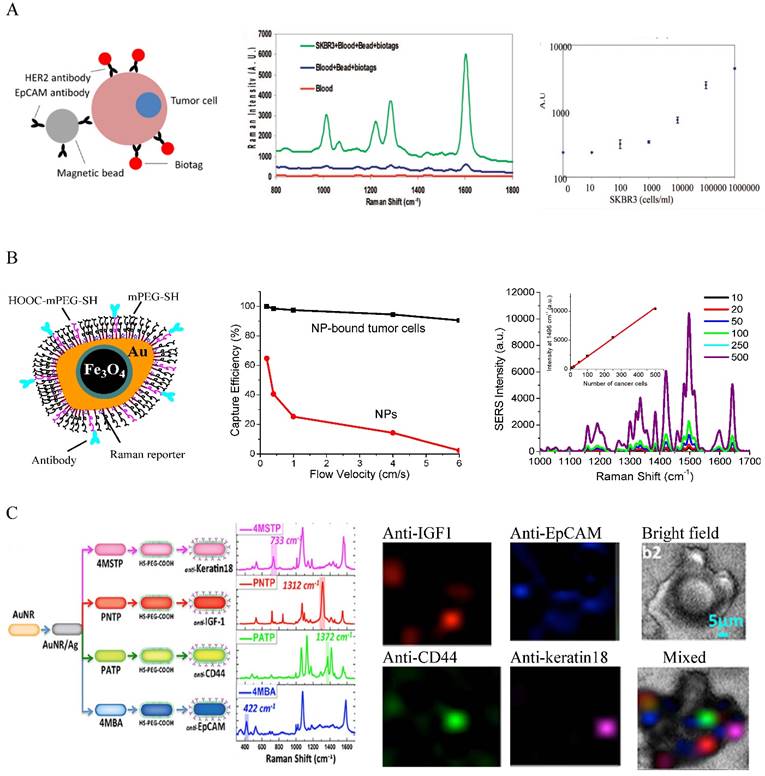
Combining isolation techniques, Au NPs have been widely used to detect CTCs based on their optical properties. A major detection method is SERS spectroscopy. SERS is known to be ultrasensitive, with sensitivity down to single molecule and single particle level due to the electromagnetic and chemical enhancements [48, 49]. Different from fluorescence spectroscopy, SERS gives sharp and fingerprinting signals. SERS bands are 10-100 times narrower than fluorescence peaks. Driven by these advantages, a new type of optical label, SERS NPs has been developed by coating Au NPs with Raman reporters (generally organic dyes with highly delocalized electrons) [50-52]. The SERS NPs are typically stabilized with methoxy polyethylene glycol thiol (mPEG-SH) with heterofunctional PEG moiety for ligand conjugation. Other molecules such as reductive bovine serum (rBSA) can also be used to ensure the biocompatibility and ligand conjugation [53]. The potential of SERS NPs for CTC detection was initially demonstrated by Sha et al. in 2008 [54] (Figure 2A). In this work, 50 nm commercial available SERS Au NPs (Nanoplex biotags) were linked with human epidermal growth factor receptor 2 (HER2) antibodies to target CTC-mimic SKBR3 breast cancer cells. Immunomagnetic enrichment with anti-EpCAM conjugated magnetic beads was used to isolate and enrich CTCs. The SERS signals are linearly proportional to the number of tumor cells, giving quantitative measurement with LOD of 10 tumor cells per mL of blood. A later strategy was to magnetically enrich the labeled cells in a tube under a flow condition followed by SERS detection [55]. The flow condition can be facilely translated into microfluidic modality for single cell analysis. Polymer membrane has also been used to capture and purify CTCs by size exclusion before SERS interrogation [56].
A major challenge in CTC detection is the interference from the large background WBCs that are difficult to be isolated from CTCs by centrifugation due to their similar densities and sizes. The use of antibodies as the recognition ligands often leads to high false positives as antibodies have nonspecific binding to WBCs. To address this challenge, Nie and co-authors used small peptide, epidermal growth factor (EGF) to recognize CTCs [57]. In their studies, CTCs were purified and enriched by density gradient centrifugation. Using QSY21 as the Raman tag and EGF as the targeting ligand, they have demonstrated that EGFR-positive head and neck cancer cells can be detected in the presence of WBCs, with LOD of 5 to 50 cancer cells per mL of blood. They have also demonstrated for the first time the clinical potential of the SERS method for CTC detection using blood samples from head and neck cancer patients. They showed that CTCs were detected in 17 out of 19 patients, with CTC numbers ranging from 1 to 720. Although the method is very promising for clinical use, it requires time consuming density gradient centrifugation for CTC separation. An advanced platform developed by our group is to integrate magnetic isolation and SERS detection with a single particle integrity, iron oxide-gold (IO-Au) core-shell NP [58] (Figure 2B). Given the dual functions and robust surface modification with pegylation, the IO-Au NPs can directly bind to CTCs in whole blood, allowing CTCs being isolated by one-step magnetic separation and subsequent on-line direction with SERS. This simple method is quick (2-h assay) and simple (no blood pretreatment). To ensure high SERS activities, we used anisotropic IO-Au nanoovals (NOVs) as the SERS substrates as anisotropic NPs have better SERS activities than spherical counterparts due to their high curvature surfaces such as particle edges and corners [59]. Using duplex targeting with anti-EpCAM and HER2 antibodies in combination with a capillary flow system, we have showed that the IO-Au NOVs can capture 90% SKBR3 cancer cells without significant interference from free IO-Au NOVs. The particles can directly bind to cancer cells in human whole blood, isolate and detect them in a single step, offering an outstanding LOD of 1-2 cells/mL of blood. SERS NPs also exhibit excellent multiplexicity. As SERS signals give a multispectral feature with extremely sharp peaks, each Raman reporter gives a unique spectral pattern. Using the distinctive peak from each reporter, multiple targets can be detected simultaneously under a single laser excitation. The SERS spectrum can be converted to images, with peak intensity yielding material concentration and peak position yielding material structure. This allows multicolor imaging of CTCs, which has been recently demonstrated by Nima et al. using silver (Ag)-coated Au NRs [60] (Figure 2C). Using four different Raman reporters, the NRs can image breast cancer cells based on expression of insulin growth factor 1 (IGF-1), anti-EpCAM, anti-CD 44, and anti-keratin 18 in the presence of WBCs. The Raman imaging offers protein tomography of multiple markers on a single cell, which is important to understand the biology of CTCs.
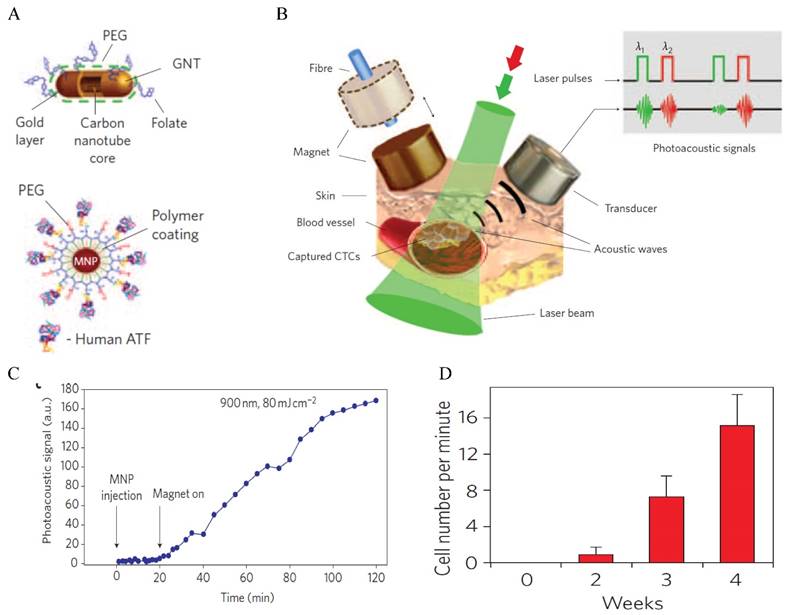
Another optical approach to detect CTCs is photoacoustic tomography (PAT), pioneered by Zharov and colleagues [61-73]. PAT combines the advantages of high contrast in optical imaging and high resolution in ultrasonics to produce super depth high resolution optical imaging [74]. In PAT detection, light from pulsed laser sources is absorbed and converted to heat that leads to transient thermoelastic expansion and wideband ultrasonic waves. The sound waves are detected and analyzed to produce images. Due to the high light absorption efficiency, Au NPs have been shown excellent PAT contrast agent for cancer imaging [75]. The NIR-absorbing properties of rod-shaped Au NPs provide excellent in vivo imaging capabilities. Imaging and detection of CTCs in vivo are important because they can monitor CTC level in real-time and detect CTCs in the entire blood volume of the body [69]. A groundbreaking work using PAT for in vivo CTC detection was conducted by Zharov and co-workers using a xenograft mouse model [64] (Figure 3). In this work, breast tumor cells circulating in mouse ear vein are magnetically enriched with 10 nm IO (Fe2O3) NPs functionalized with the amino-terminal fragment (ATF) of the urokinase plasminogen activator (UPA). As IO NPs are not strong photoacoustic agent (LOD ~ 720 NPs), gold coated carbon nanotubes (GNT) (LOD ~ 35 GNTs) were used as a second agent to boost photoacoustic signals. By converting the photoacoustic signals into cell number, they were able to monitor CTC level, with CTC rate correlating to the stage of tumor. For clinical applications, in vivo detection is still challenging because of the fast clearance of the imaging agents in the bloodstream by the reticuloendothelial system and high risk of nonspecific binding to non-tumor cells in the blood.
Au NPs can also be used to detect CTCs based on the electrocatalytic properties on hydrogen formation in the hydrogen evolution reaction (HER) [76, 77]. The Au NPs provide free electroactive sites to the protons in the acidic medium that are reduced to hydrogen by applying an adequate potential [78]. Thus, higher current is generated in the cyclic voltammogram from a solution containing Au NPs compared to control solution. When CTCs are labeled with Au NPs, the amplitude of the current in an acidic medium directly reflects the concentration of CTCs [76, 77]. This electrochemical method can detect ~ 4400 cells in suspension. By combining CTC capture to the sensing electrode with EpCAM-linked magnetic beads, the LOD was dramatically improved to 160 cells. Au NPs have also been used to directly detect CTCs in blood flow using the X-ray imaging technique [79]. By grafting Au NPs with 6-thioguanine, Jung et al. showed that they can selectively incorporate Au NPs into cancer cells but not blood cells. Because of the high Au density, cancer cells moving in the blood can be detected consecutively using the high-resolution synchrotron X-ray micro-imaging. Au NPs have also been used to detect CTCs using mass spectrometry [80-83]. Mass spectrometry has the advantages of good sensitivity, low sample requirement, and wide detection range. For example, Chiu et al. used pulsed laser desorption/ionization mass spectrometry to detect CTCs though the analysis of laser irradiation-induced Au cluster ions. It allows selective detection of MCF7 cells in blood samples at abundances as low as 10 cells.
SERS-based detection and molecular imaging of CTCs. (A) SERS Au NPs for the detection of CTCs in human whole blood. Left: Schematic of the cellular binding of antibody-conjugated SERS Au NPs (for detection) and magnetic beads (for isolation). Middle: SERS spectra of SKBR3 breast cancer cells in blood in comparison to controls. Right: Dose-response curve of SKBR3 cells in blood. Reprinted with permission from ref 54. Copyright (2008) American Chemical Society. (B) IO-Au core-shell SERS NPs for dual CTC isolation and detection. Left: Schematic of antibody-conjugated anisotropic IO-Au core-shell SERS NPs. Middle: Capture efficiencies of IO-Au core-shell NPs and SKBR3 breast cancer cells labeled with IO-Au core-shell NPs. Right: Detection of SKBR3 cells in blood with anti-HER2 and anti-EpCAM conjugated IO-Au core-shell SERS NPs. Reprinted with permission from ref 58. Copyright (2014) Future Science Group. (C) Multicolor Au-Ag core-shell SERS NRs for multiplexed CTC detection and imaging. Left: Schematic of the preparation of 4 color Au-Ag core-shell SERS NRs and their SERS spectra. Right: Raman imaging of MCF7 cells in the presence of white blood cells with the 4 color Au-Ag core-shell SERS NRs. Reprinted by permission from Macmillan Publishers Ltd: [Science Reports] (ref [60]), copyright (2014).
In vivo photoacoustic detection of CTCs with GNTs. (A) Schematic of CTC targeted GNTs (for detection) and MNPs (for enrichment). (B) Schematic of the enrichment and detection setup. (C) Photoacoustic signals from CTCs in abdominal vessels at week 1 after tumor development with and without magnetic enrichment. (D) CTC rates in mouse ear vein at different time after tumor development. Reprinted by permission from Macmillan Publishers Ltd: [Nature Nanotechnology] (ref [64]), copyright (2009).
The aforementioned detection methods are based on CTC antigen expressions. These cytometric methods allow morphological identification and downstream analyses as cells are not damaged. However, the major drawback is the lack of universal protein marker for CTC analysis. The commonly used EpCAM has heterogeneous expressions in cancer cells and its downregulation has been correlated with CTCs in peripheral blood [84]. The cytokeratin antibodies also bind to macrophages, hematopoietic cell precursors and plasma cells [85]. These limitations can be overcome by nucleic acid-based methods that detect (epi)genetic alterations specific for cancer. There are 103 to 104 copies of target RNA per CTC. Consequently, detecting RNA in CTCs can greatly enhance detection capability. Oligonucleotide-functionalized Au NPs, pioneered by Mirkin and co-workers, have been well established to detect target RNA/DNA via hybridization [86]. An example to use oligonucleotide-functionalized Au NPs to detect RNA in CTCs was demonstrated by Sioss et al. who designed a nanowire-resonator array sensor for signal readout [87]. In this method, Au NPs were bound to RNAs that were immobilized in advance on the sensor via hybridization. The Au NPs added mass to the sensor and shifted the resonance frequency of the sensor. By measuring the resonance frequency shift, the authors detected a nucleic acid prostate cancer marker, PCA3 RNA. Based on the RNA measurements and volumes used, they estimated the LOD to be 1 CTC/10 mL blood. The sensors showed high specificity, allowing single nucleotide mismatch discrimination. Due to the large surface area, Au NPs can be used as a scaffold to amplify signal production by increasing molecular binding events. An example was shown by Zhang et al. who used oligonucleotide-functionalized Au NPs to amply the binding of horseradish peroxidases (HRP) enzyme. The HRP was used to activate tyramine-biotin binding to biotin-electron rich protein for labeling with streptavidin - quantum dots (QDs) [88]. Combining a microfluidic bead capture platform, this amplification strategy led to a 1000-fold increase in detecting carcinoembryonic antigen (CEA) gene fragments compared to off-chip test. As low as 5 fM isolated CEA DNA targets from HT29 colorectal cancer cells was detected, which allowed the detection of one HT29 cell in 1 mL of blood.
Au NPs can modify the emission of fluorophores in proximity through dipole-dipole interactions. This effect depends on the particle size, shape, particle-fluorophore distance, the orientation of the fluorophore with respect to the particle, and the overlap of the emission of the molecule with the particle's absorption spectrum [18]. They can quench emission from fluorophores via non-radiative processes including Förster resonant energy transfer (FRET), electron transfer, quenching collisions, decreasing radiative decays, re-absorbing the emitted light, and chemical reactions that change the ground state of the molecules [89]. At larger distance (5 to 20 nm away), Au NPs can enhance the fluorescence by providing an external field for the fluorescence excitation and increasing the radiative decay rate of the molecule [90]. By designing a metal-fluorescence probe, analytes can be detected by their competitive binding to the particle that releases the fluorophore and thus the emission [91]. Based this detection strategy, Mirkin and co-workers developed an Au nanoFlare probe consisting of prehybridized target oligonucleotide and fluorescently labeled reporter nucleotide to detect gene markers in CTCs [92]. When the target sequence was complementary to the probe sequence, the target bound to the probe sequence, kicked off the fluorescently labeled reporter DNA, and thus restored the fluorescence of the reporter to read out the target DNA. Using this probe, they demonstrated the detection of epithelial-mesenchymal transition (EMT) marker genes (Twist, Vimentin, Fibronection, and E-cadherin) from model metastatic breast cancer cells in human blood with up to 99% fidelity. The probes were able to detect mRNAs of these genes in live cells at the single cell level. This is the first genetic-based approach for detecting, isolating and characterizing live cancer cells from blood and may provide new opportunities for CTC studies and management.
Circulating Vesicles
Extracellular vesicles (EVs) including EXOs and MVs are membrane-bound vesicles released into the extracellular environment by a spectrum of cell types, with EXOs derived from multivesicular bodies and MVs from plasma membrane [93-95]. They carry molecular constitutes of their originating cells [96-99] and represent an important mode of intercellular communication by horizontal transfer of proteins and nucleic acids between cells [98-102]. They were reported to play important roles during cancer initiation and progression [103-106], including activation of normal epithelial cells to form tumors [103] and conversion of indolent cancer cells to active cancer cells [108]. They have been found in various body fluids such as blood, urine, saliva, and cerebrospinal fluid [109-112]. Generally, patients with advanced cancer have higher amount of EVs than healthy subjects [113]. Thus, EVs have emerged as a potential new class of cancer biomarkers for clinical diagnostics and treatment monitoring [114-117].
EXOs and MVs in plasma require purification and enrichment before detection and characterization. The most commonly used method is ultracentrifugation that involves a series of differential centrifugations followed by high speed centrifugation to pellet the membrane bound vesicles [118]. Density gradient centrifugation, filtration, and immunomagnetic isolation are also common methods to capture and concentrate vesicles [119]. After isolation, vesicles are generally characterized by nanoparticle tracking analysis (NTA) to determine the size and concentration and by scanning electron microscope (SEM) to examine the morphology. The detection and molecular analysis of circulating vesicles are challenging due to their small size (40-100 nm for EXOs and 100-1000 nm for MVs) and heterogeneous compositions. They cannot be examined by conventional optical imaging. Nor can they be analyzed by flow cytometry when their sizes are smaller than 300 nm [120]. Conventional characterization methods such as Western blotting and ELISA require lengthy processes and concentrated EVs from large volume of samples (3 mL plasma and 300 mL cell culture media) [121, 122], making them impractical in a typical clinical setting.
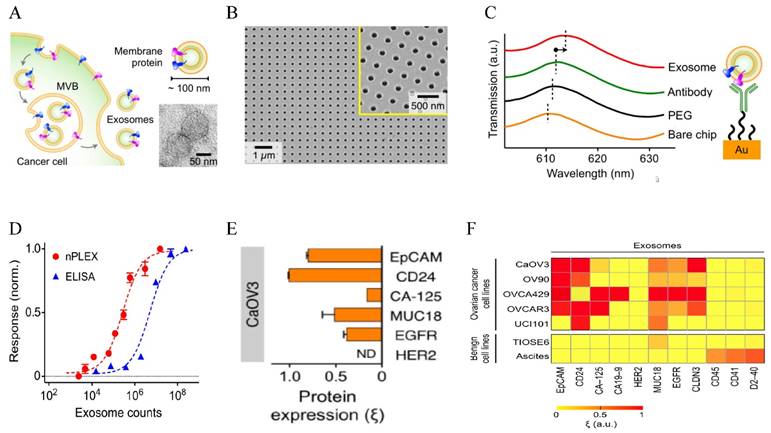
Au-based nanostructures provide a promising sensing platform based on the LSPR shift in response to the changes of the environment. The LSPR wavelength is linearly proportional to the refractive index of the surrounding medium based on the Drude model, with larger Au NPs giving higher sensitivity (the relative shift of the LSPR wavelength with respect to the refractive index change of the surroundings) [123]. As a matter of fact, this refractive index sensing based on SPR shift from solid Au surface has been a standard commercialized approach to detect molecular binding events and various chemical and biological species [124]. The LSPR-based sensing gives better linearity and higher resolution [125] and can reach single molecule sensitivity using appropriate amplification mechanisms [126] or single particle spectroscopy [127]. Using the LSPR sensing strategy, Lee and co-workers designed a novel sensor for EXO detection and profiling, named nanoplasmonic exosome (nPLEX) assay [19] (Figure 4). The sensor was made of periodic Au nanohole arrays on a glass substrate, with the hole diameter of 200 nm and periodicity of 450 nm. Each array was functionalized with target-specific antibodies to capture EXOs. The LSPR peak red shifted upon EXO binding and the spectral shifts or intensity changes were proportional to the surface protein levels on EXOs. Using EXO surface marker CD63, they have established that the assay had a LOD of 670 aM. This sensitivity was 104 times higher than western blot and 102 higher than ELISA. The assay allows high throughput (over 50 markers) quantification of a panel of surface proteins in a multiplexed fashion by combining the nanohole array with a miniaturized multichannel chip. Studies on clinical samples showed that EXOs can be used as a marker for diagnostics and treatment monitoring for ovarian cancer based on EXO expressions of EpCAM and CD24. They further showed that the additional EXO labeling with Au NPs improved signals by 20 % and with Au nanostars (NSTs) by 300 %. Given the high sensitivity and high throughput, this nPLEX assay has strong potential for diagnosis and real-time treatment monitoring for a variety types of cancers.
The LSPR peak is also sensitive to the interparticle distance. Individual Au NPs exhibit red color with LSPR around 520 nm. When Au NPs are brought close to each other, the solution changes color to blue, with LSPR red shifting and broadening because of plasmonic coupling. The fractional spectral shift decays exponentially with the interparticle distance. The decay length is about 0.1 in units of the particle size for different particle size, shape, metal type and medium dielectric constant, a behavior called “universal scaling” [128]. This colorimetric property has been widely used to design various sensors for molecular and cellular detections [129]. Recently, Maiolo et al. demonstrated colorimetric detection of EVs based on NP aggregation and the corresponding spectral changes when Au NPs were bound onto EVs via nonspecific electrostatic interactions [130]. EVs as low as 35 fM were detected. Although the sensitivity of this colorimetric method is lower than the aforementioned nPLEX assay, this method is simpler, cheaper and quicker, permitting its use in resource-limited clinical settings for cancer screening and real-time treatment evaluation.
Majority EV detection techniques are based on bulk analysis, unable to assess EXO and MV subpopulations. In fact, a single cell can release multiple subtypes of EVs. Therefore, techniques capable of analyzing single vesicles are highly valuable. Braeckmans and co-authors exploited the feasibility of single EXO chemical analysis using the SERS technology [131]. The 10 nm Au NPs capped with cationic 4-dimethylaminopyridine (DMAP) were incubated with purified EXOs from B16F10 melanoma cells to allow the adsorption of Au NPs (~ 600 Au NP/EXO) onto EXOs via electrostatic interactions. The Au NP-coated EXOs were plated on quartz slide and SERS spectra were recorded using a confocal Raman microscope to obtain signals from single vesicles. Due to the dense packing of Au NPs, the exosomal biomolecules including lipids, proteins, carbohydrates, and even nucleic acids could be detected. These fingerprinting SERS signals are different from those by the RBC control. Of course, the DMAP capping materials also gave strong SERS signals, but they are distinct from EXO signals. Further, the authors could identify EXOs from B16F10 cells in the presence of EXOs from RBCs based on chemical analysis. Thus, this method shows the strong potential to discriminate vesicles from different cellular origins for diagnostic implications.
Lateral flow immunoassay (LFIA) is a great tool for cost-effective on-site detection [132]. It uses the same detection mechanism as ELISA except that it runs in a dipstick format and the solid phase is nitrocellulose membrane instead of a plastic well. It is a one-step and rapid assay, suitable for point-of-care (POC) detection. Recently, Oliveira-Rodriguez et al. applied LFIA to detect EVs using Au NPs as the optical labels [133]. In this study, EXOs from human metastatic melanoma cells in conditioned media were purified by ultracentrifugation. Anti-CD9 and anti-CD81 were used as the capture antibodies and 40 nm Au NPs were used as the detection agent. Line intensities from bound Au NPs were recorded by scanning the images. The optical density of Au NPs was measured to give colorimetric detection. The whole procedure took only 15 min. The method reaches LOD of 8.54 x 105 EXO/μL. The clinical potential of this method was tested with plasma and urine samples from melanoma patients. Their later studies showed that Au NPs gave better sensitivity and linear detection range than two other labels-carbon black NPs (CB) and magnetic nanoparticles (MNPs) [134]. They further developed the technique into a multiplexed assay by using different antibodies on different test lines. This allows the detection of a broad range of EVs based on their surface protein tomography.
LSPR-based nanoplasmonic exosome sensor (nPLEX) for exosome detection and molecular profiling. (A) Schematic of exosome biogenesis. (B) Scanning electron micrograph of the periodic Au nanoholes in the sensor. (C) Absorption spectra of the Au nanoholes after binding with PEG, antibody, and exosomes, respectively. (D) Comparison of the sensitivity of nPLEX with ELISA. (E) Quantitative molecular profiling of surface markers on exosomes from CaOV3 ovarian cancer cells. (F) Molecular profiling of putative ovarian cancer markers (EpCAM, CD24, CA19-9, CLDN3, CA-125, MUC18, EGFR, HER2), immune host cell markers (CD41, CD45) and a mesothelial marker (D2-40) on exosomes from different cell lines. Reprinted by permission from Macmillan Publishers Ltd: [Nature Biotechnology] (ref [19]), copyright (2014).
Circulating Nucleic Acids (CNAs)
Dying tumor cells release small fragments (50 to 250 bp) of their DNA into the bloodstream [135]. These fragments, first found in cancer patients in 1977 [136], are called cell-free ctDNA. They carry genomic and epigenomic alterations identical to those of tumor tissues and discriminate from normal cell-free DNA [137]. ctDNA is broadly applicable, specific and sensitive biomarker that can be used for wide range of research and clinical purposes, including tumor genotyping, early cancer detection, patient prognosis, therapy evaluation, and minimal residual disease monitoring [138-145]. Thus, ctDNA is becoming a new generation of biomarker for cancer assessment. Another type of CNAs is circulating RNA. Since early discoveries of tumor-associated messenger RNA (mRNA) in the plasma of cancer patients in 1990s [146-148], circulating RNAs including mRNA, microRNA (miRNA), and other non-coding RNAs have been repeatedly found in the blood plasma of patients with a variety of types of cancers. miRNA level in serum and plasma is more reproducible, stable, and consistent compared to other types of RNAs [149]. Similar to ctDNA, circulating RNAs have been successfully evaluated in many types of cancers and emerged as promising biomarkers for cancer diagnosis, staging, prognosis and treatment monitoring [150-154].
Due to the relatively low amount, CNAs need to be isolated and enriched from plasma or serum before analysis. Commercial isolation kits are available, including QIAamp Circulating Nucleic Acid Kit, Invitrogen™ MagMAX™ Cell-Free DNA Isolation Kit, ZR serum DNA kit, PME free-circulating DNA Extraction Kit, Plasma/Serum Cell-Free Circulating DNA Purification Mini Kit, NucleoSpin Plasma XS, FitAmp Plasma/Serum DNA Isolation Kit, FitAmp Plasma/Serum DNA Isolation Kit, and Chemagen's circulating NA kit. After extraction and concentration, ctDNA is typically analyzed by PCR, fluorescence, or spectrophotometry-based approaches, with recent technologies of droplet digital PCR (ddPCR), deep sequencing, and whole genome sequencing [144, 145]. The gold standard RNA detection method is quantitative real-time (qRT)-PCR. Broad gene expression platforms either based on quantitative PCR (qPCR) or hybridization techniques exist and commercially RNA detection assays are also available [155]. However, the PCR-based methods require costly and complex procedures and often give high false positive signals. The use of nanomaterials can simplify the procedure, be more cost-friendly and more specific.
Au NP-based DNA assays were pioneered by Mirkin and colleagues who first developed a colorimetric method based on DNA-mediated Au NP assembly in 1997 [156]. They designed two oligonucleotide-functionalized Au NP probes with sequences complementary to different segments of the targeted polynucleotides. When the two oligonucleotide - Au NP conjugates bound to the target via hybridization, they were cross-linked and aggregated, inducing spectroscopic shift and red-to-blue color change that were detected by UV-Vis spectroscopy or naked eye. This method is very specific, being able to distinguish nucleotide sequences with single base mismatch [156-158]. Later, they developed a bio-barcode method with PCR-like sensitivity that uses two-component oligonucleotide-modified Au NPs for signal recognition and single-component oligonucleotide-modified magnetic beads for separation [159]. Detection of amplified barcode DNA was achieved with a chip-based scanometric method. After that, extensive development of Au NP-based DNA/RNA assays have been reported, which can be mainly categorized into colorimetric methods based on cross-linking aggregation, non-cross-linking aggregation, sandwich assays (e.g. microarrays), fluorescence, LSPR, SERS, and electrochemical methods [160]. However, majority techniques have been only tested on commercially synthesized oligonucleotides.
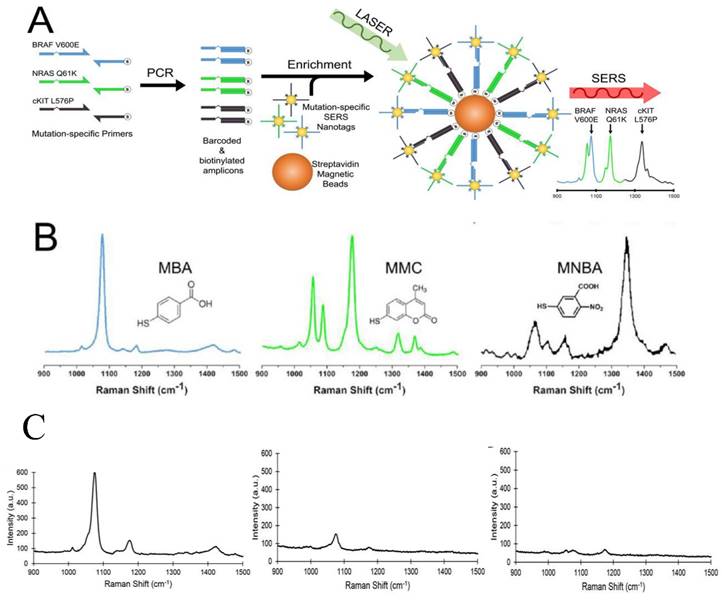
The analysis of ctDNA is challenging because ctDNA is a small fraction (often <1%) within the high background levels of wild-type cell-free DNA. Recently, Wee et al. reported a SERS-based method in conjunction with PCR to detect as few as 10 mutant alleles from a background of 10,000 wild type sequences (0.1%) (Figure 5) [161]. Three primers specific to three clinically important DNA point mutations BRAF V600E, NRAS Q61K, and c-Kit L576P were used to amplify tumor DNA. Amplicons were then tagged with BRAF V600E, NRAS Q61K, and c-Kit L576P-specific SERS nanotags and enriched using magnetic beads. These mutations are encoded with three Raman reporters, 4-Mercaptobenzoic acid (MBA), 2,7-mercapto-4-methylcoumarin (MMC) or 4-mercapto-3-nitrobenzoic acid (MNBA). The presence of the targeted mutation was detected by the encoded SERS nanotags using Raman spectroscopy. Using less than 5 ng of genomic DNA, the method could accurately genotype cell lines and ctDNA in serum samples from melanoma patients. This multiplexed method has assay time comparable to current qPCR-based methods, with potential sensitivity to ddPCR.
Existing methods for ctDNA detection are restricted to genetic mutations. A dual genetic and epigenetic detection method for ctDNA was reported by Nguyen and Sim based on the LSPR sensing [162]. The authors made peptide nucleic acid (PNA)-functionalized Au NPs to target two bio-signatures of ctDNA: mutations at two hot-spots E542K and E545K of the PIK3CA gene and methylation. PNA is advantageous over DNA because it does not have electrostatic interference with the negatively charged phosphate backbone of target DNA and its complex with mismatched DNA is more destabilizing than that in DNA/DNA. The PNA-functionalized Au NPs were immobilized on a glass substrate to capture and enrich the 69-bp PIK3CA ctDNA. Binding of ctDNA to the PNA-Au NPs were observed by the red shift of the dark field light scattering LSPR peak of single Au NPs. At the optimal temperature of 62oC, they observed LSPR shift of 4.3 nm after incubation with 200 fM synthetic ctDNA in contrast to 0.1 nm for the normal circulating DNA, indicating the detection of the signature mutations on ctDNA. Subsequently, 20 nm anti-mc immunogold was exploited as the methylation detectors that further red shifted the LSPR from 4.3 nm to 11.4 due to plasmonic coupling with the PNA-Au NPs, leading to detection down to 50 fM. Thus, hot-spot mutations and epigenetic changes on the ctDNA were detected in one-step by the nanoplasmonic biosensor, opening a new approach for detecting ctDNA biosignatures at high sensitivity and specificity.
Combining the high selectivity of biochemical recognition and high sensitivity of electrochemical detection, electrochemical biosensors provide an attractive means to analyze the content of a biological sample [163]. Nanomaterials have been well used in electrochemical biosensors to improve capture efficiency via better interactions with analytes than solid substrate [164]. Using Au NRs in conjunction with graphene oxide (GO), Azimzadeh et al. reported an electrochemical nanobiosensor to detect miR-155 in plasma, an important oncogenic circulating miRNA [165]. Au NRs were decorated on GO sheet on the surface of the glassy carbon electrode (working electrode) and then functionalized with thiolated single stranded (SS) probe targeting miR-155, a circulating miRNA. After target binding via hybridization, the sensor was treated with electrochemical indictor oracet blue (OB). OB intercalated with the SS-miRNA hybrid and its reduction signal was measured by the differential pulse voltammetry (DPV) method. Hybridization with the target miRNA induced much more decrease in the current than the control. The current was linearly related to the concentration of target miRNA, with an outstanding LOD of 0.6 fM. The sensor also had extremely high specificity. It discriminated complementary target miRNA from single base mismatched miRNA. Using the spiking miRNA in healthy human blood, the sensor showed a ~100 % recovery via direct detection of the target in plasma without the need of sample separation and signal amplification. Due to the high sensitivity, high specificity and easy operation, the method has strong promise for early cancer detection.
Multiplexed detection of melanoma DNA mutatons in ctDNA with SERS nanotags. (A) Schematic of the multiplexed SERS assay in combination with PCR for detection of BRAF V600E, NRAS Q61K, and c-Kit L576P mutations in ctDNA. (B) Molecular structure and SERS spectra of the SERS nanotags. (C) Typical SERS spectra of 3 ctDNA serum samples from melanoma patients with highly positive (left), low (middle), and negative (right) BRAF V600 E mutations. Cited from ref [161].
Techniques capable of multiplexed detection are highly desirable because they allow simultaneous measurements of different RNAs and can look for many different types of cancer. Using the aggregation-based colorimetric properties of Au NPs in combination with DNA hybridization chain reaction (HCR), Rana et al. demonstrated a reprogrammable and multiplexed detection method for circulating miRNAs [166]. They immobilized a target specific 36-mer capture-DNA on Au NPs. The target oncomiRs hybridized with the DNA probe on Au NPs to form an RNA/DNA heteroduplex with a sticky RNA end. A specific initiator was then added to bind to the RNA, forming an Au NP assembly with a sticky DNA to open up the first hairpin. The first hairpin initiated the opening of second hairpin and started the HCR process. As a result, orthogonal DNA polymers were formed on Au NPs, which prevented the aggregation of the particles from magnesium ion (Mg2+)-induced aggregation and maintained the red color of individual Au NPs. In the absence of target oncomiRs, the Au NPs aggregated with the addition of Mg2+ inducing a color change from red to blue. Using this method, they showed simultaneous detection of three oncomiRs, miR-10b, miR-21, and miR-141 by simply changing the DNA probe sequence. As low as 20 fmol of each miR was visually detected without using any instrument. Compared to the colorimetric method without HCR [167], this method gives similar sensitivity but can detect multiple miRNAs at the same time, making it promising for clinical applications.
Circulating Proteins
Proteins circulating in the blood have been investigated intensively in proteomics in searching for biomarkers for cancer diagnosis. Protein biomarkers are typically released from cells or organs and they are characteristic of pathophysiologic and physiologic conditions. Cancer patients often display elevated level of proteins that are correlated to metastasis, prognosis and therapeutic response. Many proteins have been identified as potential serum markers for a variety of types of cancers [168-177]. Some proteins have been routinely used in clinical practice to help get information about the presence or absent of disease as well as its evolution, including prostate-specific antigen (PSA) for prostate cancer, cancer antigen 125 (CA 125) for ovarian cancer, alpha-fetoprotein (AFP) for liver cancer, CA19.9 for gastric/pancreatic cancer, carcinoembryonic antigen (CEA) for colorectal cancer, and cancer antigen 15.3 (CA15.3)/CA27.29 for breast cancer [178]. As part of the immunodefense against tumor, autoantibodies in cancer patients such as immunoglobulin G (IgG) and immunoglobulin E (IgE) are usually higher than noncancerous patients. Thus, autoantibodies in serum are also biomarkers for cancer diagnosis. Classic methods for protein biomarker analysis include western blot, ELISA, RIA, and mass spectrometry [179]. However, these methods are either insensitive for early detection or technically complicated and time-consuming. For example, the commercially available immunoassays have a typical LOD at the picomolar level [8]. However, the blood concentrations of protein biomarkers associated with early state cancers range from 10-16 to 10-12 M [7]. Thus, it is very important to develop new assays with easy operation and high sensitivity.
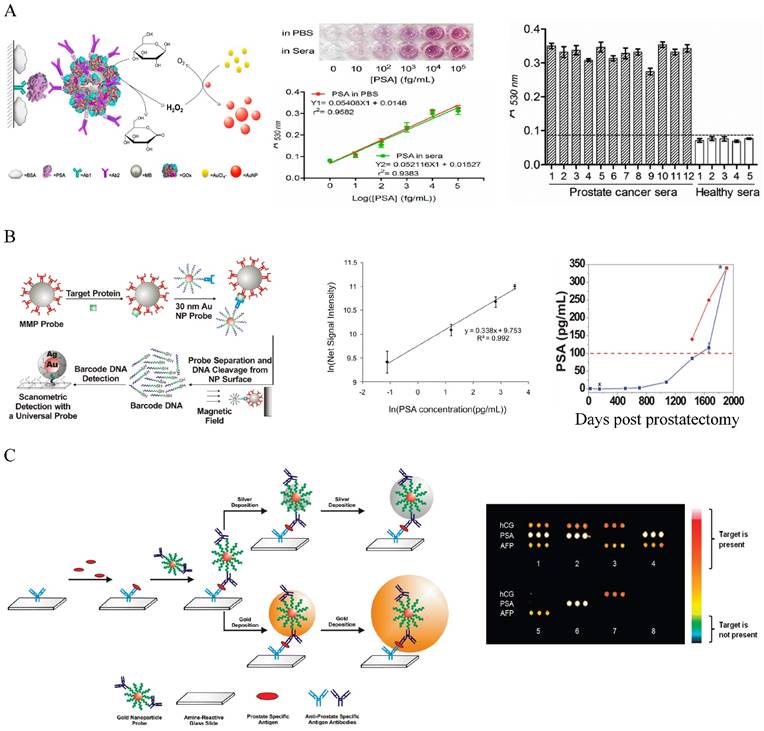
The most widely used method for protein marker detection is colorimetric immunoassays that use antibody-antigen interactions with enzyme or optical labels for signal readout on the detection antibody [180]. Au NPs can enhance the traditional colorimetric assays by increasing sensor-analyte interactions. By coating ELISA plate with a layer of Au NPs, the LOD for CEA detection was improved by more than 5 times due to the enhanced protein adsorption on the Au NP-coated plate [181]. Au NPs have been used as the scaffold for the detection probes to amplify signal readout in the classic enzyme-catalyzed assays, but these approaches have moderate sensitivity (ng to pg/mL level) [172-185]. Colorimetric methods based on particle aggregation from individual particles allow protein detection with naked eyes with comparable sensitivity to the conventional ELISA [186-188]. Using the simple UV-Vis spectroscopy to determine the concentrations of Au NPs in the presence and absence of proteins can detect protein markers in the picomolar range [189]. A high sensitivity Au NP-amplified ELISA assay was developed by Rica and Stevens in 2012 by linking the biocatalytic cycle of the enzyme catalase to the reduction of Au ions to obtain colored Au NPs of characteristic tonality [190]. More specifically, in the absence of the analyte, the rate of Au precursor (AuCl4-) reduction by hydrogen peroxide (H2O2) was high, leading to non-aggregated red Au NPs. In the presence of analytes, the enzyme catalase that was conjugated on a secondary antibody consumed H2O2, causing AuCl4- reduction at low rate. This led to the formation of aggregated Au NPs in blue color with ill-defined morphology. The LOD was as low as 10-18 g/mL for PSA in whole serum. Although the sensitivity is high, quantitative detection is difficult because of the narrow linear detection range. A high sensitivity and high throughput ELISA-like approach with wide linear range was reported by Chen and co-workers who linked glucose oxidase (GOx)-catalyzed amplification to the growth of small colorless Au NPs to large colored Au NPs as the signal readout [191] (Figure 6A). In this method, GOx was conjugated with detection antibody to the immunomagnetic beads that sandwiched PSA with capture antibody on a 96-well polystyrene plate. Glucose was added to generate H2O2 via the GOx catalyzed oxidation. In the presence of H2O2 and AuCl4-, the small colorless Au NPs turned into red. The absorbance at 530 nm was measured to quantify the amount of produced large Au NPs. The method gave an unprecedentedly wide linear range between 10 and 105 fg/mL PSA, with LOD of 4.6 fg/mL (93 aM). This is 104 times more sensitive than the HRP-based commercial ELISA kit (0.21 ng/mL). Clinical sample tests showed that the method successfully detected serum PSA in patients that were undetectable by the conventional ELISA. If commercialized, this assay will provide a revolutionary platform for low cost and POC detection of serum protein cancer makers in both resource-rich and resource-limited settings.
Another remarkable application is the use of Au NPs to develop bio-barcode assays without enzymatic amplification that can be 4 - 6 orders of more sensitive than ELISA [20, 192-195]. The center of the biobarcode assays, which has been commercialized, is to use Au NPs conjugated with both barcode oligonucleotides and target-specific antibodies for signal amplification (Figure 6B) [195]. Proteins in solution are first captured with immunomagnetic beads via magnetic separation. Au NP conjugates are added to sandwich the proteins with the immunomagnetic beads. After magnetic separation, the barcode oligonucleotides are released by dehybridization and detected by the chip-based scanometric assay that takes the advantage of Au-NP catalyzed silver enhancement. This method can detect PSA in PBS at attomolar (10-18 M) level, as originally demonstrated by the Mirkin group [20]. The assay has been automated and tested later on clinical samples that showed its capability to follow serum PSA from recurrent prostate cancer patients under prostatectomy that could not be detected by commercial assays [195]. Multiplexed detection has been achieved by varying the detection antibodies, barcode oligonucleotides as well as capture nucleotides on chip stripes [193]. Au NPs have also been used to develop microarray assays by taking the advantages of the multiplexicity of protein microarray technology and the high sensitivity of scanometric detection (Figure 6C) [196]. In this type of assays, the antibody microarray is first fabricated by spotting of different antibody solutions into different micro-sized pins on a glass slide. Proteins captured on the microarray are then sandwiched by the Au NP probes and directly detected by the scanometric method. This method can detect multiplexed proteins at low picomolar concentrations in buffer or in diluted serum. Although the method gives lower sensitivity than the barcode method, they are simple, fast, high throughput, and require small amount of agents.
Au NP-based colorimetric immunoassays for the detection of cancer serum proteins. (A) Au NP-based ELISA. Left: Schematic illustration of the quantitative immunoassay based on GOx-catalyzed growth of Au NPs (5 nm in diameter). Middle: Quantitative detection of PSA in different concentrations in PBS and sera samples with naked-dye (top) and microplater reader (bottom). Right: Detection of PA in human sera using the Au NP-based assay. Cited from ref 191. (B) Au NP-based biobarcode assay. Left: Schematic illustration of the Au NP-based biobarcode assay. Middle: Scanometric detection of serum PSA with different concentrations. Right: Detection of serum PSA in a recurrent patient after radical prostatectomy. The dotted redline indicates the undeteable PSA region by the coventional commerical immunoassays (Abbott IMx assay). Data collected by the commercial assay was shown in red dot and by the bio-barcode assay in blue dots. Cited from ref 195. (C) Au NP -based microarray assay. Left: Schematic illustration of the Au NP -based microarray assay showing scanometric detection with silver deposition enhancement and gold deposition enhancent. Right: Scanometric detection of three protein cancer markers HCG, PA and AFP in buffer in eight different samples after two gold depositions. Reprinted with permission from ref [196]. Copyright (2009) American Chemical Society.
Fluorescence enhanced protein microarray on Au/Au film. (A) Scanning electron micrograph of the Au/Au film (Au island nanostructures deposited on solid Au film). (B) Extinction spectrum of the Au/Au film overlapping with the excitations of Cy5 and IR800. (C) Fluorescence intensity maps showing about 100 x stronger fluorescent signals than the glass controls for both Cy5 and IR800. (D) Calibration curve for CEA detection with the Au/Au film compared to that with the glass control. Inset shows the schematic illustration of the sandwich assay. (E) Fluorescence intensity maps of autoantigen microarrays on the Au/Au film (μArray/Au) probed with human serum containing autoantibodies. (F) Intensity heatmap comparing the mean fluorescence intensities of the autoantigens on μArray/Au in comparison to those on nitrocellulose and glass controls. Reprinted by permission from Macmillan Publishers Ltd: [Nature Communications] (ref [197]), copyright (2011).
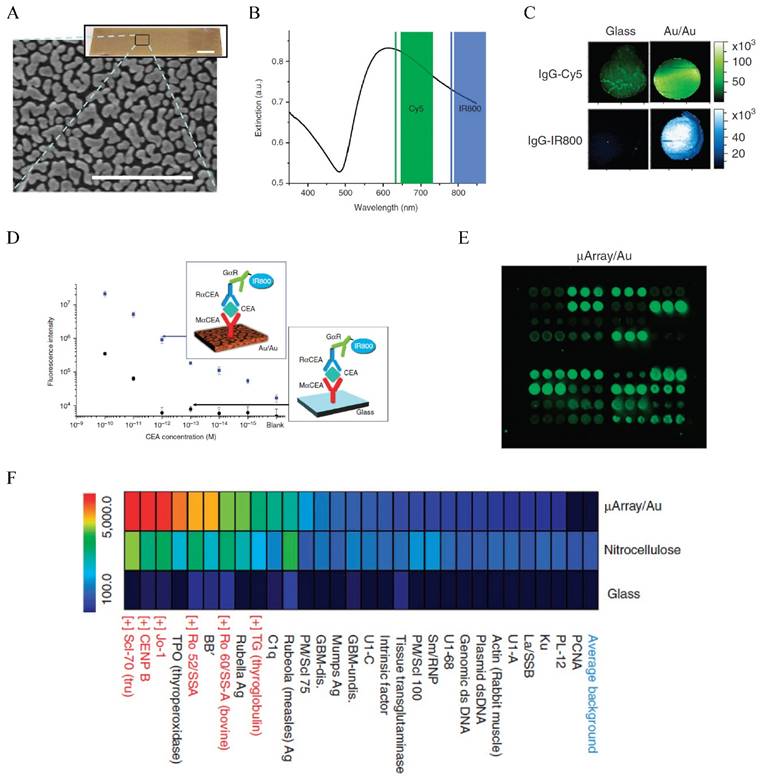
The effect of Au NPs on the fluorescence properties of adjacent dye or particles has been used to detect miRNA in CTCs as mentioned earlier. This detection mechanism has also been used to detect serum protein markers, as demonstrated by Dai and co-workers who used fluorescence enhancement by nanostructured Au on Au film to develop a new generation protein microarray assay (Figure 7) [197]. The Au/Au film on glass, prepared by reduction of AuCl4- on a glass slide, gave strong absorption in the NIR region overlapping with the excitation of NIR fluorophores such as Cy5 and IR800. Thus, the fluorescence of these molecules was enhanced up to 100-fold. Using a microarray printing robot, the Au/Au substrate was made into a microarray with 400 μm spots functionalized with anti-CEA antibodies. Using a sandwich assay with IR800 tagged detection antibody, they demonstrated detection of CEA in whole serum down to femtomolar level in contrast to 100 pM by the glass substrate, extending the dynamic range by three orders of magnitude towards the femtomolar regime. Using this method, they monitored the CEA level in a LS174T xenograft mouse model and showed that the CEA serum concentration was correlated to the tumor volume. They further made a high throughput autoantigen array to screen autoantibodies in human sera, which offered higher positive signals owing to the fluorescence enhancement than the nitrocellulose and glass substrate controls. This method presents new opportunities in proteomic research and cancer diagnosis. Another example is presented by Liu et al. who used cadmium telluride QDs as the fluorescence donor and Au NPs as the acceptor to develop a homogeneous immunoassay for CEA detection [198]. In the presence of CEA, the antibody-protein binding made the QDs and Au NPs close enough that the fluorescence of QDs was quenched. The fluorescence variation was linearly correlated to the CEA concentration. Although the performance of the assay (LOD 0.3 ng/mL and linear range 1-100 ng/mL) is moderate, it is extremely simple and can be extended to detect a wide range of biomarkers.
SPR is the gold standard method for label-free optical biosensing. However, conventional SPR has several concerns including limited sensitivity, low throughput and difficult miniaturization. A variety of signal enhancement strategies have been developed, with LOD for serum proteins reaching down to zeptomolar level [199]. One of the strategies is to use Au NP-tagged antibody to sandwich protein with an Au film support to enhance the SPR response [200-203]. A distinguished example was demonstrated by Sim et al. who showed that the use of antibody-conjugated Au NRs enhanced SPR signals by 108 compared to conventional SPR measurement, which allowed the detection of IgE at the attomolar level [202]. To improve throughput, Couture et al. developed an approach to allow SPR reading in a standard 96-well plate [204]. This was achieved by making an Au nanohole array on a glass wafer with a photolithographic process. The Au nanohole arrays, 720 nm in diameter and 1200 nm in periodicity, served as a multiwall plate reader for plasmonic sensing of tens of samples at the same time. Rather than using Au NPs to enhance SPR response of solid Au film, many studies use the LSPR shift of Au NPs as signal readout to avoid the use of expensive instrument [205]. However, the sensitivity of conventional LSPR assays is very limited although researchers have used a variety of anisotropic Au NPs to improve the refractive index sensitivity. Single particle spectroscopy provides a great opportunity for ultrasensitive sensing as it can reach detection at single molecule level [206]. An example for circulating protein marker detection is the single nanobiosensor demonstrated by Sim and co-workers who used LSPR response from single Au NRs to detect PSA binding [207]. Anti-PSA antibody conjugated Au NRs with aspect ratio of 2.75 were immobilized onto a glass slide as the sensing platform. Upon PSA binding, the light scattering LSPR wavelength of Au NRs, collected by dark field microspectroscopy, red shifted with distinguishable 2.9 nm shift in responding to 111 aM PSA. In their later work, they found the sensitivity depended on the aspect ratio of Au NRs, with the optimal Au NRs with aspect ratio of 3.5 giving LOD of 1 aM [208]. This single nanobiosensor enables simple and label-free high sensitivity detection of targeted protein markers. Although single particle sensing is highly sensitive, it is challenging as it is extremely difficult to make Au NRs of identical sizes. To develop highly sensitive LSPR sensing, Stevens and co-authors linked the biocatalytic cycle of GOx enzyme to the crystal growth of Au NSTs to amplify signal readout [209]. PSA was sandwiched between primary antibody and immune Au NSTs and purified by centrifugation. GOx was linked on the detection antibody that catalyzed the reduction of O2 to H2O2. At low concentrations of GOx, the self-nucleation of Ag atoms was slow. Instead, Ag ions (Ag+) were reduced by H2O2 on Au NSTs to form Ag-Au core-shell NSTs. This led to blue-shift of the LSPR wavelength of Au NSTs. At high concentrations of GOx, the self-nucleation rate of Ag atoms was high, which led to the formation of Ag NPs rather than Ag-coated Au NSTs. Under this condition, the LSPR of Au NSTs exhibited a small LSPR shift. This signal-generation step induced an inverse sensitivity to the protein, with LOD of 10-18 g/mL (4 x 10-20 M) and linear range of 10-18 g/mL to 10-13 g/mL for spiked PSA in whole serum.
Due to their high sensitivity, good photostability, large working range, and excellent multiplexicity, SERS has become another major method for serum protein marker detection [210-220]. In typical SERS-based assays, protein markers are sandwiched between antibody-conjugated SERS Au NPs and an antibody-coated surface either a two-dimensional substrate (typically glass slides coated with thin Au film) [213-216] or magnetic beads [217-220]. The nontargeted biomolecules in serum samples are readily separated by washing or magnetic separation. Studies using spherical SERS Au NPs and hollow Au nanospheres (HGNs) showed a LOD of 1 pg/mL, comparable to commercial immunoassays [213, 217]. Anisotropic Au NPs such as NSTs have been lately used to improve the detection sensitivity because of their much stronger SERS enhancement on the high curvature tips and edges [214, 215]. A unique assay with high sensitivity was reported by Li et al. who used SERS Au NSTs (malachite green isothiocyanate as the Raman reporter) as the detection agents and Au triangle nanoarray as the substrate [215]. Due to the plasmonic field coupling, the SERS Au NSTs and the nanoarray substrate formed a strong and confined electromagnetic field in the 3D space. This dramatically decreased the LOD down to 7 fg/mL (~0.3 fM) for the human IgG protein in the buffer solution that they tested. The assay was tested with clinical blood samples from breast cancer patients for biomarker vascular endothelial growth factor (VEGF) to diagnose tumor-associated angiogenesis. By designing a functional array on the substrate for different markers, the SERS-based immunoassay can be used for multiplexed detection. For example, Li et al. constructed a 3 x 7 array on an Au-coated glass slide with parafilm defined wells, which allowed simultaneous detection of CA15.3, CA27.29 and CEA in different concentrations [214]. With significantly lower LOD than conventional ELISA, this assay has also been evaluated with real clinical samples from breast cancer patients, which all showed significantly higher concentration of CA15.3, CA27.29 and CEA than healthy serum samples.
Different from the commonly used immunoassay methods with SERS NPs, Chen and co-workers used Au NPs for label-free SERS detection that can probe cancer specific biomolecular changes rather than quantification [221, 222]. The procedure is extremely simple, just mixing Au NPs directly with patient blood serum samples. This method can not only detect proteins, but also nucleic acid and saccharide in serum. Findings have been reported that colorectal cancer patients have higher amount of nucleic acid and lower amount of saccharide and proteins than healthy subjects. Combining with principal components analysis (PCA) and linear discriminant analysis (LDA), the method yielded a diagnostic sensitivity of 97% and specificity of 100% for colorectal cancer [221]. The method could differentiate nasopharyngeal cancer at different states, with 83.5% diagnostic accuracy for T1 stage and 93.3% for T2-T4 stage. It is known that Ag has higher SERS activity than Au due to less plasmon damping by interband electron transitions. Thus, Zheng and co-workers used Ag NPs as the SERS substrates to detect nasopharyngeal, gastric, and colorectal cancers based on the biochemical SERS signatures of serum blood samples [223-225]. These studies indicate that the label-free SERS method has great potential for non-invasive detection of a variety types of cancers.
As mentioned earlier, nanostructure-based electrochemical sensors have become popular because they offer improved biomolecular interactions that can increase not only dynamic range of measurements but also detection sensitivity. This is primarily achieved by making nanostructured electrode surface [226-232]. For example, Arkan et al. deposited Au NPs on a multiwall carbon nanotube (MWCNT)-ionic liquid electrode, which offered the LOD of HER2 in serum samples around 7 ng/mL using the electrochemical impedance spectroscopy (EIS) [228]. Feng et al. deposited Au NPs onto an Au electrode and detected both CEA and AFP by sandwiching the proteins with MWCNTs functionalized with anti-CEA and anti-AFP antibodies respectively [229]. This AuNPs@MWCNT method offered a linear range of 0.01 to 60 ng/mL and a LOD of 3.0 pg/mL for CEA and 4.5 pg/mL for AFP. A high sensitivity method was recently reported by Presnova et al. who deposited 5 nm Au NPs on silicon nanowire field-effect transistor (NW-FET) [230]. The NW-FET functionalized exhibited extraordinary performance, with LOD of 23 fg/mL for PSA and working range of 23 fg/mL to 500 ng/mL due to the combination of high sensitivity of the nanowire and strong signal enhancement by Au NPs. A new electrochemical strategy was demonstrated by Ilkhani et al. who took the advantages of high specificity of the sandwich assay, high efficiency of magnetic separation, and high sensitivity of electrochemical detection [231]. In these studies, EGFR proteins were sandwiched between aptamer-coated magnetic beads and anti-EGFR conjugated Au NPs. The extent of the complexation, thus the level of EGFR in serum was monitored by differential pulse voltammetry of Au NPs after dissolution with an acidic medium. The method offered a LOD of 50 pg/mL. Clinical efficacy has been tested by monitoring EGFR level in serum of breast cancer patients.
A typical heterogeneous immunoassay involves multiple steps of incubation, washing cycles, signal amplification and reading. This takes hours to days to complete. Huo and co-workers reported a one-step, wash-free, and homogenous immunoassay using DLS as the detection method [233]. When Au NRs with capture antibody, Au NPs with detection antibody, and PSA were mixed together, the PSA linked the antibody-conjugated Au NRs and Au NPs together via antibody-antigen bindings, inducing particle aggregation and thus the increase of particles' size. The size change of the particle solution, measured by DLS, was linearly proportional to the PSA concentration, with LOD reaching down to 0.02 pM. The method only took 30 min before DLS measurement. Later, they used this simple DLS method to detect cancer from the blood with nonfunctionalized citrate- capped Au NPs [234]. The hypothesis is that the protein profile in the blood of cancer patients is different from healthy donors, thus the molecular composition of the protein corona formed on the Au NP surface may differ between cancerous and noncancerous human blood. Therefore, cancer-specific antigens should be in the protein corona layer that can be probed with antibodies. Rabbit anti-human IgG was added to probe the antibodies. The multiplexed binding of the anti-human IgG onto Au NPs induced particle aggregation and subsequent size changes that were measured by DLS. Using this method, the authors showed that the amount of IgG in the blood serum of prostate cancer patients was significantly higher than noncancerous controls. The test gave a 90-95-% specificity and 50% sensitivity in detecting early stage prostate cancer. A different approach in developing simple-to-operate immunoassay with high sensitivity was recently reported by Tamayo et al. [235]. CEA or PSA was first captured on an array of silicon microcantilevers driven by a piezoelectric actuator. Then, 100 nm Au NPs linked with a second antibody were applied to sandwich the protein with the cantilever. The Au NPs served as a mass and plasmonic label. Due to the mass loading, the attachment of the Au NPs onto the cantilever caused the cantilever vibration at a lower frequency that was measured by a laser beam deflection method. The resulting downshift of the resonance frequency was proportional to both the added mass and the amount of protein targets. Meanwhile, the cantilever served as an optical cavity that boosted the scattering signal from the Au NPs due to plasmonic coupling. In the absence of proteins, the scattering signal from the cantilever was negligible. Increased scattering signals were observed in the presence of CEA or PSA due to the binding of Au NPs. This optomechanical method offered a LOD of 0.1 fg/mL in serum for both CEA and PSA. which was at least seven orders of magnitude lower than that achieved in routine clinical practice.
Conclusions and Future Perspectives
Rapid advancements in cancer medicine have identified a variety of cancer-related circulating biomarkers, classified as circulating tumor cells, circulating vesicles, circulating nucleic acids, and circulating proteins. These circulating cancer biomarkers offer the possibility of noninvasive cancer screening, early detection, patient prognosis, and enable monitoring of tumor growth and treatment response. Therefore, the development of effective detection tools is of great importance. Undoubtedly, traditional analytical methods such as flow cytometry, ELISA, DNA and protein arrays have made great contributions to push this field forward. However, they have moderate sensitivity, unable to provide early cancer detection and screening. Many emerging technologies with unprecedented sensitivity have been detected for each class of these biomarkers with nanotechnology playing a key role. The Au NPs have become one of the major types of nanoplatforms and their utilizations have led to many different assays based on different detection strategies including LSPR, colorimetry, SERS, fluorescence, photoacoustics, and electrochemistry. Depending on the target and the signal readout mechanism, each of these methods has different applications. For example, SERS have been used for detection and molecular analysis of all four markers. However, the LSPR-based sensing is useful for detection of exosomes, DNA, and protein markers, but not for large size CTCs due to their sensitivity to molecular binding events.
Despite the great progress in existing Au NP-based platforms, new technologies need to be developed to address several needs. Current CTC technologies are focusing on detection. However, clinical significance of CTCs remains to be determined. To enable CTCs to be more clinically informative, effective technologies for CTC molecular characterizations are highly desirable. The SERS Au NPs are promising for high sensitivity, high specificity and high throughput protein profiling on single CTCs. Research has shown the ability to separate the spectral fingerprints of up to 10 different types of SERS NPs [236]. In contrast, such high throughput analysis on a single cell using fluorescence-based methods is extremely challenging. Combining with portable devices such as microfluidics, the SERS technology would improve our understanding of CTCs and cancer progression, which may make enormous impact on basic cancer research and clinical patient management. Although exosomes and microvesicles have shown strong clinical potential, their biology remains largely elusive. Understanding their genetic and proteomic knowledge is critical to accelerate their clinical applications. The nPLEX assay developed by Lee and co-workers represents a novel platform for high throughput analysis of surface protein markers [19]. However, the nPLEX chip is costly and technically complicated, involving highly specialized ion-beam milling and soft lithography. This makes it impractical for resource-limited clinical settings. New technologies need to be simple, low cost, and rapid without deteriorating sensitivity and throughput. In addition, next generation technologies should also emphasize on detection and analysis at single vesicle level, which will significantly improve our understanding on circulating vesicles and may lead to new discovery that is inherently missing with bulk methods. For the nucleic acid detection, current Au NP-based methods are mainly based on proof-of-concept studies on synthetic samples. Validation studies on clinical samples need to be reinforced to accelerate their translation. A variety of Au-based methods with unsurpassed sensitivity have been tested on serum protein markers, but they have not been translated into clinical practices. Extensive developments are needed before clinical applications to generate large scale and reproductive Au NP probes with precise control of size, shape, composition, and surface chemistry. In addition, transformation into portable devices is also needed for simple and femtomolar diagnosis.
A combination of different types of cancer makers is often needed to give accurate assessment of various aspects of cancer. Mao et al. demonstrated an approach for simultaneous detection of nucleic acid and protein with DNA and antibody functionalized Au NPs in combination of a lateral flow device [237]. However, the sensitivity is moderate, with LOD of 0.5 nM DNA and 2 ng/mL IgG. Thus, new technologies for simultaneous detection of two or more different types of markers with high sensitivity are essential in the development of simple and POC diagnosis. A remaining challenge for biomarker analysis is the sample preparation due to the rarity of circulating biomarkers and the complicated blood environment. Nanomaterials with dual isolation and detection functions such as IO-Au core-shell NPs provide one great opportunity to address this challenge, but validations on clinical samples are insufficient. Another way is to combine Au NPs with microfluidics devices. Microfluidics provides a venue for producing highly integrated system that can process clinical samples in closed architectures to minimize sample contamination and loss. It enables user-friendly automation on a single device, reducing human intervention and operational errors. The microfluidic chip is very important for enriching low freqency bioamrkers to reduce false signals. It has become a major approach for isolation and concentration of circulating biomarkers especially CTCs based on size, deformity or affinity mechanisms. Thus, Au NP-based microfluidic platforms will be sought-after in the next five to ten years.
Acknowledgements
We acknowledge the financial support from the National Cancer Institute, a part of the National Institutes of Health (Grant No. 1R15 CA 195509-01).
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Kim ES, Hirsh V, Mok T. et al. Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): a randomised phase III trial. Lancet. 2008;372:1809-18
2. Brock G, Castellanos-Rizaldos E, Hu L, Coticchia C, Skog J. Liquid biopsy for cancer screening, patient stratification and monitoring. Transl Cancer Res. 2015;4:280-90
3. Diaz Jr LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579-86
4. Miller MC, Doyle GV, Terstappen LWMM. Significance of circulating tumor cells detected by the CellSearch systemin patients with metastatic breast colorectal and prostate cancer. J Oncol. 2010;2010:617421
5. Diehl F, Li M, Dressman D. et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci. USA. 2005;102:16368-73
6. Diehl F, Schmidt K, Choti MA. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985-90
7. Rissin DM, Kan CW, Campbell TG. et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28:595-9
8. Rosi NL, Mirkin CA. Nanostructures in Biodiagnostics. Chem Rev. 2005;105:1547-56
9. Swierczewska M, Liu G, Lee S, Chen X. High-sensitivity nanosensors for biomarker detection. Chem Soc Rev. 2012;41:2641-55
10. Link S, El-Sayed MA. Shape and size dependence of radiative, non-radiative and photothermal properties of gold nanocrystals. Int Rev Phys Chem. 2000;19:409-53
11. Kelly KL, Coronado E, Zhao LL, Schatz GC. The optical properties of metal nanoparticles: the influence of size, shape, and dielectric environment. J Phys Chem B. 2003;107:668-77
12. Willets KA, Van Duyne RP. Localized surface plasmon resonance spectroscopy and sensing. Annu Rev Phys Chem. 2007;58:267-97
13. Li N, Zhao P, Astruc D. Anosotropic gold nanoparticles: synthesis, properties, applications, and toxicity. Angew Chem Int Ed. 2014;53:1756-89
14. Hurst SJ, Lytton-Jean AKR, Mirkin CA. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal Chem. 2006;78:8313-18
15. Orendorff CJ, Murphy CJ. Quantitation of metal content in the silver-assisted growth of gold nanorods. J Phys Chem B. 2006;110:3990-4
16. Xia Y, Halas NJ. Shape-controlled synthesis and surface plasmonic properties of metallic nanostructures. MRS Bulletin. 2005;30:338-48
17. Kneipp K, Kneipp H, Itzkan H, Dasari RR, Feld MS. Surface-enhanced Raman scattering and biophysics. J Phys Condens Matter. 2002;14:R597-624
18. Gersten J, Nitzan A. Spectroscopic properties of molecules interacting with small dielectric particles. J Chem Phys. 1981;75:1139-52
19. Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H. Label-free detection and molecular profiling of exosomes with a nanoplasmonic sensor. Nat Biotechnol. 2014;32:490-5
20. Nam JM, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884-6
21. Bardhan R, Surbhio L, Joshi A, Halas NJ. Theranostic nanoshells: from probe design to imaging and treatment of cancer. Acc Chem Res. 2011;44:936-46
22. Dreaden EC, Alkilany AM, Huang X, Murphy CJ, El-Sayed MA. The golden age: gold nanoparticles for biomedicine. Chem Soc Rev. 2012;41:2740-79
23. Saha K, Agasti SS, Kim C, Li X, Rotello VM. Gold nanoparticles in chemical and biological Sensing. Chem Rev. 2012;112:2739-79
24. Zhou W, Gao X, Liu D, Chen X. Gold Nanoparticles for in vitro diagnostics. Chem Rev. 2015;115:10575-636
25. Yang X, Yang M, Pang B, Vara M, Xia Y. Gold nanomaterials at work in biomedicine. Chem Rev. 2015;115:10410-88
26. Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275-92
27. Liberko M, Kolostova K, Bobek V. Essentials of circulating tumor cells for clinical research and practice. Crit Rev Oncol Hematol. 2013;88:338-56
28. Alix-Panabières CKP. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. 2013;59:110-8
29. Mocellin S, Hoon D, Ambrosi A, Nitti D, Rossi CR. The prognostic value of circulating tumor cells in patients with melanoma: a systematic review and meta-analysis. Clin Cancer Res. 2006;12:4605-13
30. De Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LWWM, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302-9
31. Krebs MG, Sloane R, Priest L, Lancashire L, Hou JM, Greystoke A, Ward TH, Ferraldeschi R, Hughes A, Clack G. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol. 2011;29:1556-63
32. Hiltermann TJN, Pore MM, van den Berg A. et al. Circulating tumor cells in small-cell lung cancer: A predictive and prognostic factor. Ann Oncol. 2012;23:2937-42
33. Hartkopf AD, Wagner P, Wallwiener D, Fehm T, Rothmund R. Changing levels of circulating tumor cells in monitoring chemotherapy response in patients with metastatic breast cancer. Anticancer Res. 2011;31:979-84
34. Miyamoto DT, Sequist LV, Lee RJ. Circulating tumour cells-monitoring treatment response in prostate cancer. Nat Rev Clin Oncol. 2014;11:401-12
35. Smerage JB, Barlow WE, Hortobagyi GN. et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J Clin Oncol. 2014;32:3483-89
36. Hüsemann Y, Geigl JB, Schubert F. et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58-68
37. Arya SK, Lim B, Rahman ARA. Enrichment, detection and clinical significance of circulating tumor cells. Lab Chip. 2013;13:1995-2027
38. Liberti PA, Rao CG, Terstappen LWMM. Optimization of ferro#uids and protocols for the enrichment of breast tumor cells in blood. J Magn Magn Mater. 2001;225:301-7
39. Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AGJ, Uhr JW, Terstappen LWMM. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897-904
40. Van der Auwera I, Peeters D, Benoy IH, Elst HJ, Van Laere SJ, Prove A, Maes H, Huget P, van Dam P, Vermeulen PB, Dirix LY. Circulating tumour cell detection: a direct comparison between the CellSearch system, the AdnaTest and CK-19/mammaglobin RT-PCR in patients with metastatic breast cancer. British J Cancer. 2010;102:276-84
41. Andreopoulou E, Yang LY, Rangel KM, Reuben JM, Hsu L, Krishnamurthy S, Valero V, Fritsche HA, Cristofanilli M. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect™ versus Veridex CellSearch™ system. Int J Cancer. 2012;130:1590-7
42. Brökera P, Lücke K, Perpeet M, Gronewold TMA. A nanostructured SAW chip-based biosensor detecting cancer cells. Sens Actuator B. 2012;165:1-6
43. Park GS, Kwon H, Kwak DW. et al. Full surface embedding of gold clusters on silicon nanowires for efficient capture and photothermal therapy of circulating tumor cells. Nano Lett. 2012;12:1638-42
44. Wang Y, Zhou F, Liu X, Yuan L, Li D, Wang Y, Chen H. Aptamer-modified micro/nanostructured surfaces: efficient capture of Ramos cells in serum environment. ACS Appl Mater Interfaces. 2013;5:3816-23
45. Sheng W, Chen T, Tan W, Fan ZH. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano. 2013;7:7067-76
46. Lv SW, Liu Y, Xie M, Wang J, Yan XW, Li Z, Dong WG, Huang WH. Near-infrared light-responsive hydrogel for specific recognition and photothermal site-release of circulating tumor cells. ACS Nano. 2016;10:6201-10
47. Feng Y, Sun F, Chen L, Lei J, Ju H. Ratiometric electrochemiluminescence detection of circulating tumor cells and cell-surface glycans. J Electroanaly Chem. 2016
48. Nie S, Emory SR. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275:1102-6
49. Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari RR, Feld MS. Single Molecule Detection Using Surface-Enhanced Raman Scattering (SERS). Phys Rev Lett. 1997;78:1667-70
50. Doering WE, Piotti ME, Natan MJ, Freeman RG. SERS as a foundation for nanoscale, optically detected biological labels. Adv Mater. 2007;19:3100-8
51. Sha MY, Xu H, Penn SG, Cromer R. SERS nanoparticles: a new optical detection modality for cancer diagnosis. Nanomedicine. 2007;2:725-34
52. Lane LA, Qian X, Nie S. SERS nanoparticles in medicine: from label-free detection to spectroscopic tagging. Chem Rev. 2015;115:10489-529
53. Wu X, Luo L, Yang S, Ma X, Li Y, Dong C, Tian Y, Zhang L, Shen Z, Wu A. Improved SERS nanoparticles for direct detection of circulating tumor cells in the blood. ACS Appl Mater Interfaces. 2015;7:9965-71
54. Sha MY, Xu HX, Natan MJ, Cromer R. Surface-enhanced Raman scattering tags for rapid and homogeneous detection of circulating tumor cells in the presence of human whole blood. J Am Chem Soc. 2008;130:17214-5
55. Shi W, Paproski RJ, Moore R, Zemp R. Detection of circulating tumor cells using targeted surface-enhanced Raman scattering nanoparticles and magnetic enrichment. J Biomed Optics. 2014;19:056014
56. Zhang P, Zhang R, Gao M, Zhang X. Novel nitrocellulose membrane substrate for efficient analysis of circulating tumor cells coupled with surface-enhanced Raman scattering imaging. ACS Appl Mater Interfaces. 2014;6:370-6
57. Wang X, Qian XM, Beitler JJ, Chen ZG, Khuri FR, Lewis MM, Shin HJC, Nie SM, Shin DM. Detection of circulating tumor cells in human peripheral blood using surface-enhanced Raman scattering nanoparticles. Cancer Res. 2011;71:1526-32
58. Bhana S, Chaffin E, Wang Y, Mishra SR, Huang X. Capture and detection of cancer cells in whole blood with magnetic-optical nanoovals. Nanomedicine(Lond). 2014;9:593-606
59. Hao E, Schatz G, Hupp J. Synthesis and optical properties of anisotropic metal nanoparticles. J Fluor. 2004;14:331-41
60. Nima ZA, Mahmood M, Xu Y. et al. Circulating tumor cell identification by functionalized silver-gold nanorods with multicolor, super-enhanced SERS and photothermal resonances. Sci Rep. 2014;4:4752
61. Zharov V, Galanzha E, Shashkov E, Khlebtsov N, Tuchin V. In vivo photoacoustic flow cytometry for monitoring circulating singe cancer cells and contrast agents. Opt Lett. 2006;31:3623-5
62. Holan SH, Viator JA. Automated wavelet denoising of photoacoustic signals for circulating melanoma cell detection and burn image reconstruction. Phys Med Biol. 2008;53:N227-36
63. Galanzha EI, Shashkov EV, Spring PM, Suen JY, Zharov VP. In vivo, noninvasive, label-free detection and eradication of circulating metastatic melanoma cells using two-color photoacoustic flow cytometry with a diode laser. Cancer Res. 2009;69:7926-34
64. Galanzha EI, Shashkov EV, Kelly T, Kim JW, Yang L, Zharov VP. In vivo magnetic enrichment and multiplex photoacoustic detection of circulating tumour cells. Nat Nanotechnol. 2009;4:855-60
65. Galanzha EI, Kim JW, Zharov VP. Nanotechnology-based molecular photoacoustic and photothermal flow cytometry platform for in-vivo detection and killing of circulating cancer stem cells. J Biophoton. 2009;2:725-35
66. Viator JA, Gupta S, Goldschmidt BS, Bhattacharyya K, Kannan R, Shukla R, Dale PS, Boote E, Katti K. Gold nanoparticle mediated detection of prostate cancer cells using photoacoustic flowmetry with optical reflectance. J Biomed Nanotechnol. 2010;6:187-91
67. Zharov VP. Ultrasharp nonlinear photothermal and photoacoustic resonances and holes beyond the spectral limit. Nat Photon. 2011;5:110-6
68. Bhattacharyya K, Goldschmidt BS, Hannink M, Alexander S, Viator JA. Gold nanoparticle mediated detection of circulating cancer cells. Clin Lab Med. 2012;32:89-101
69. Galanzha EIVPZ. Circulating tumor cell detection and capture by photoacoustic flow cytometry in vivo and ex vivo. Cancer. 2013;5:1691-738
70. Hu X, Wei CW, Xia J, Pelivanov I, O'Donnell M, Gao X. Trapping and photoacoustic detection of CTCs at the single cell per milliliter level with magneto-optical coupled nanoparticles. Small. 2013;9:2043-52
71. Nedosekin DA, Sarimollaoglu M, Galanzha EI, Sawant R, Torchilin VP, Verkhusha VV, Ma J, Frank MH, Biris AS, Zharov VP. Synergy of photoacoustic and fluorescence flow cytometry of circulating cells with negative and positive contrasts. J Biophotonics. 2013;6:425-34
72. Nedosekin DA, Juratli MA, Sarimollaoglu M, Moore CL, Rusch NJ, Smeltzer MS, Zharov VP, Galanzha EI. Photoacoustic and photothermal detection of circulating tumor cells, bacteria and nanoparticles in cerebrospinal fluid in vivo and ex vivo. J Biophotonics. 2013;6:523-533
73. Juratli M, Sarimollaoglu M, Siegel E, Nedosekin DA, Galanzha EI, Suen JY, Zharov VP. Real-time monitoring of circulating tumor cell release during tumor manipulation using in vivo photoacoustic and fluorescent flow cytometry. Head Neck. 2014;36:1207-15
74. Yao J, Wang LV. Photoacoustic tomography: fundamentals, advances and prospects. Contrast Media Mol Imaging. 2011;6:332-45
75. Li W, Chen X. Gold nanoparticles for photoacoustic imaging. Nanomedicine (Lond.). 2015;10:299-320
76. Costa MM, Escosura-Muñiz A, Nogués C, Barrios L, Ibáñez E, A Merkoçi. Simple monitoring of cancer cells using nanoparticles. Nano Lett. 2012;12:4164-71
77. Costa MM, Escosura-Muñiz A, Nogués C, Barrios L, Ibáñez E, MerkoçI A. Detection of circulating cancer cells using electrocatalytic gold nanoparticles. Small. 2012;8:3605-12
78. Costa MM, Escosura-Muñiz A, Merkoçi A. Electrochemical quantification of gold nanoparticles based on their catalytic properties toward hydrogen formation: Application in magnetoimmunoassays. Eectrochem Commun. 2010;12:1501-4
79. Jung SY, Ahn S, Seo E, Lee SJ. Detection of circulating tumor cells via an X-ray imaging technique. J Synchrotron Rad. 2013;20:324-31
80. Chiu WJ, Ling TK, Chiang HP, Lin HJ, Huang CC. Monitoring cluster ions derived from aptamer-modified gold nanofilms under laser desorption/ionization for the detection of circulating tumor cells. ACS Appl Mater Interfaces. 2015;7:8622-30
81. Zhang X, Chen B, He M, Wang H, Hu B. Gold nanoparticles labeling with hybridization chain reaction amplification strategy for the sensitive detection of HepG2 cells by inductively coupled plasma mass spectrometry. Biosens Bioelectron. 2016;86:736-40
82. Baird Z, Pirro V, Ayrton S, Hollerbach A, Hanau C, Marfurt K, Foltz M, Cooks RG, Pugia M. Tumor cell detection by mass spectrometry using signal ion emission reactive release amplification. Anal Chem. 2016;88:6971-5
83. Li X, Chen B, He M, Wang H, Xiao G, Yang B, Hu B. Simultaneous detection of MCF-7 and HepG2 cells in blood by ICP-MS with gold nanoparticles and quantum dots as elemental tags. Biosens Bioelectron. 2017;90:343-8
84. Danova M, Torchio M, Mazzini G. Isolation of rare circulating tumor cells in cancer patients: technical aspects and clinical implications. Expert Rev Mol Diagn. 2011;11:473-85
85. Borgen E, Beiske K, Trachsel S. et al. Immunocytochemical detection of isolated epithelial cells in bone marrow: non-specific staining and contribution by plasma cells directly reactive to alkaline phosphatase. J Pathol. 1998;185:427-34
86. Storhoff JJ, Mirkin CA. Programmed materials synthesis with DNA. Chem Rev. 1999;99:1849-62
87. Sioss JA, Bhiladvala R, Pan W. et al. Nanoresonator chip-based RNA sensor strategy for detection of circulating tumor tells: response using PCA3 as a prostate cancer marker. Nanomedicine. 2012;8:1017-25
88. Zhang H, Fu X, Hu J, Zhu Z. Microfluidic bead-based multienzyme-nanoparticle amplification for detection of circulating tumor cells in the blood using quantum dots labels. Anal Chim Acta. 2013;779:64-71
89. Dulkeith E, Morteani AC, Niedereichholz T, Klar TA, Feldmann J. Fluorescence quenching of dye molecules near gold nanoparticles: radiative and nonradiative effects. Phys Rev Lett. 2002;89:203002
90. Lakowicz JR. Radiative decay engineering 5: metal-enhanced fluorescence and plasmon emission. Anal Biochem. 2005;337:171-94
91. Swierczewska M, Lee S, Chen X. The design and application of fluorophore-gold nanoparticle activatable probes. Phys Chem Chem Phys. 2011;13:9929-41
92. Haloa TL, McMahonc KM, Angelonic NL. et al. NanoFlares for the detection, isolation, and culture of live tumor cells from human blood. Proc Natl Acad Sci USA. 2014;111:17104-9
93. Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci. 2000;113:3365-74
94. Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Cell. 2009;19:43-51
95. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373-83
96. Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180
97. Graner MW, Alzate O, Dechkovskaia AM, Keene JD, Sampson JH, Mitchell DA, Bigner DD. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009;23:1541-57
98. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-9
99. Skog J, Wurdinger T, van Rijn S, Meijer D, Gainche L, Sena-Esteves M, Curry Jr WT, Carter RS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and protein that promote tumor growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470-6
100. Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delive. Leukemia. 2006;20:847-56
101. Mathivanan SHJ, Simpson RJ. Exosomes: Extracellular organelles important in intercellular communication. J. Proteomics. 2010;73:1907-20
102. Lo Cicero A, Stahl PD, Raposo G. Extracellular vesicles shuffling intercellular messages: for good or for bad. Curr Opin Cell Biol. 2015;35:69-77
103. Ginestra A, La Placa MD, Saladino F, Cassarà D, Nagase H, Vittorelli ML. The amount and proteolytic content of vesicles shed by human cancer cell lines correlates with their in vitro invasiveness. Anticancer Res. 1998;18:3433-7
104. Valenti R, Huber V, Iero M, Filipazzi P, Parmiani G, Rivoltini L. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 2007;67:2912-5
105. Lee TH, D'Asti E, Magnus D, Al-Nedawi K, Meehan B, Rak J. Microvesicles as mediators of intercellular communication in cancer-the emerging science of cellular 'debris'. Semin Immunopathol. 2011;33:455-67
106. D'Souza-Schorey C, Clancy JW. Tumor-derived microvesicles: shedding light on novel microenvironment modulators and prospective cancer biomarkers. Genes Dev. 2012;26:1287-99
107. Melo SA, Sugimoto H, O'Conner JT. et al. Cancer Exosomes Perform Cell-Independent MicroRNA Biogenesis and Promote Tumorigenesis. Cancer Cell. 2014;26:707-21
108. Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619-24
109. Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879-87
110. Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101:13368-73
111. Ogawa Y, Miura Y, Harazono A. et al. Proteomic analysis of two types of exosomes in human whole saliva. Biol Pharm Bull. 2011;34:13-23
112. Vella LJ, Sharples RA, Lawson VA, Masters CL, Cappai R, Hill AF. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J Pathol. 2007;211:582-90
113. Logozzi M, De Milito A, Lugini L. et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4:e5219
114. Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267-83
115. Choi DS, Lee J, Go G, Kim YK, Gho YS. Circulating extracellular vesicles in cancer diagnosis and monitoring: an appraisal of clinical potential. Mol Diagn Ther. 2013;17:265-71
116. Santiago-Dieppa DR, Steinberg J, Gonda D, Cheung VJ, Carter BS, Chen CC. Extracellular vesicles as a platform for ''liquid biopsy'' in glioblastoma patients. Expert Rev Mol Diagn. 2014;14:819-25
117. Verma M, Lam TK, Hebert E, Divi RL. Extracellular vesicles: potential applications in cancer diagnosis, prognosis, and epidemiology. BMC Clin Pathol. 2015;15:1-9
118. Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, Simpson RJ. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293-304
119. Lobb RJ, Becker M, Wen SW, Wong CSF, Wiegmans AP, Leimgruber A, Mo¨ ller A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicle. 2015;4:27031
120. Inglis H, Norris P, Danesh A. Techniques for the analysis of extracellular vesicles using flow cytometry. J Vis Exp. 2015;97:e52484
121. Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys. 2012;1820:940-8
122. Taylor DD, Zacharias W, Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol Biol. 2011;728:235-46
123. Lee SK, El-Sayed MA. Gold and silver nanoparticles in sensing and imaging: sensitivity of plasmon response to size, shape, and metal composition. J Phys Chem B. 2006;110:19220-5
124. Homola J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 2008;108:462-93
125. Cao J, Galbraith EK, Sun T, Grattan KTV. Comparison of surface plasmon resonance and localized surface plasmon resonance-based optical fibre sensors. J Phys. 2011;307:012050
126. Chen S, Svedendahl M, Van Duyne RP, Käll M. Plasmon-enhanced colorimetric ELISA with single molecule sensitivity. Nano Lett. 2011;11:1826-30
127. Mayer KM, Hao F, Lee S, Nordlander P, Hafner JJ. single molecule immunoassay by localized surface plasmon resonance. Nanotechnology. 2010;21:255503
128. Jain PK, Huang W, El-Sayed MA. On the universal scaling behavior of the distance decay of plasmon coupling in metal nanoparticle pairs: a plasmon ruler equation. Nano Lett. 2007;7:2080-8
129. Zhang Y, Mckelvie ID, Cattrall RW, Kolev SD. Colorimetric detection based on localised surface plasmon resonance of gold nanoparticles: Merits,inherent short comings and future prospects. Talanta. 2016;152:410-22
130. Maiolo D, Paolini L, Noto GD, Zendrini A, Berti D, Bergese P, Ricotta D. Colorimetric nanoplasmonic assay to determine purity and titrate extracellular vesicles. Anal Chem. 2015;87:4168-76
131. Stremersch S, Marro M, Pinchasik B. et al. Identifi cation of individual exosome-like vesicles by surface enhanced Raman spectroscopy. Small. 2016;12:3292-301
132. Posthuma-Trumpie GA, Korf J, vanAmerongen A. Lateral flow (immuno)assay: its strengths, weaknesses, opportunities and threats. A literature survey. Anal Bioanal Chem. 2009;393:569-82
133. Oliveira-Rodríguez M, López-Cobo S, Reyburn HT. et al. Development of a rapid lateral flow immunoassay test for detection of exosomes previously enriched from cell culture medium and body fluids. J Extracell Vesicles. 2016;5:31803-13
134. Oliveira-Rodríguez M, Serrano-Pertierra E, García AC, Martín SL, Mo MY, Cernuda-Morollón E, Blanco-López MC. Point-of-care detection of extracellular vesicles: sensitivity optimization and multiple-target detection. Biosens Bioelectron. 2017;87:38-45
135. Jahr S, Hentze H, Englisch S. et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659-65
136. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646-50
137. Ignatiadis M, Dawson SJ. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol. 2014;25:2304-13
138. Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer-A survey. Biochim Biophys Acta. 2007;1775:181-232
139. Elshimali YI, Khaddour H, Sarkissyan M, Wu Y, Vadgama JV. The clinical utilization of circulating cell free DNA (CCFDNA) in blood of cancer patients. Int J Mol Sci. 2013;14:18925-58
140. Marzese DM, Hirose H, Hoon DS. Diagnostic and prognostic value of circulating tumor-related DNA in cancer patients. Expert Rev Mol Diagn. 2013;13:827-44
141. Diaz Jr LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579-86
142. Bettegowda C, Sausen M, Leary RJ. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:Ra24
143. Karachaliou N, Mayo-de-las-Casas C, Molina-Vila MA, Rosell R. Real-time liquid biopsies become a reality in cancer treatment. Ann Transl Med. 2015;3:36
144. Heitzer E, Ulz P, Geigl JB. Circulating Tumor DNA as a Liquid Biopsy for Cancer. Clin Chem. 2015;61:112-23
145. Qin Z, Ljubimov VA, Zhou C, Tong Y, Liang J. Cell-free circulating tumor DNA in cancer. Chin. J Cancer. 2016;35:36
146. Funaki NO, Tanaka J, Kasamatsu T. et al. Identification of carcinoembryonic antigen mRNA in circulating peripheral blood of pancreatic carcinoma and gastric carcinoma patients. Life Sci. 1996;59:2187-99
147. Kopreski MS, Benko FA, Kwak LW, Gocke CD. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clin Cancer Res. 1999;5:1961-5
148. Lo KW, Lo YM, Leung SF. et al. Analysis of cell-free Epstein-Barr virus associated RNA in the plasma of patients with nasopharyngeal carcinoma. Clin Chem. 1999;45:1292-4
149. Redova M, Sana J, Slaby O. Circulating miRNAs as new blood-based biomarkers for solid cancers. Future Oncol. 2013;9:387-402
150. Garcia V, Garcia JM, Pena C. et al. Free circulating mRNA in plasma from breast cancer patients and clinical outcome. Cancer Lett. 2008;263:312-20
151. March-Villalba JA, Martinez-Jabaloyas JM, Herrero MJ. et al. Cell-free circulating plasma hTERT mRNA is a useful marker for prostate cancer diagnosis and is associated with poor prognosis tumor characteristics. PLoS One. 2012;7:e43470
152. Garcia V, Garcia JM, Silva J. et al. Extracellular tumor-related mRNA in plasma of lymphoma patients and survival implications. PLoS One. 2009;4:e8173
153. Redova M, Sana J, Slaby O. Circulating miRNAs as new blood-based biomarkers for solid cancers. Future Oncol. 2013;9:1-16
154. Kishikawa T, Otsuka M, Ohno M, Yoshikawa T, Takata A, Koike K. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J Gastroenterol. 2015;21:8527-40
155. Jensen SG, Lamy P, Rasmussen MH. et al. Evaluation of two commercial global miRNA expression profiling platforms for detection of less abundant miRNAs. BMC Genomics. 2011;12:435
156. Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078-81
157. Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. One-Pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J Am Chem Soc. 1999;120:1959-64
158. Reynolds RA, Mirkin CA, Letsinger RL. Homogeneous, nanoparticle-based quantitative colorimetric detection of oligonucleotides. J Am Chem Soc. 2000;122:3795-6
159. Nam JM, Stoeva SI, Mirkin CA. Bio-bar-code-based DNA detection with PCR-like sensitivity. J Am Chem Soc. 2004;126:5932-33
160. Franco R, Pedrosa P, Carlos FF, Veigas B, Baptista PV. Gold Nanoparticles for DNA/RNA-based diagnostics. In: (ed.) Aliofkhazraei M. Handbook of Nanoparticles. Springer International Publishing. 2015:1339-64
161. Wee EJH, Wang Y, Tsao SCH, Trau M. Simple, sensitive and accurate multiplex detection of clinically important melanoma DNA mutations in circulating tumour DNA with SERS nanotags. Theranostics. 2016;6:1506-13
162. Nguyen AH, Sim SJ. Nanoplasmonic biosensor: Detection and amplification of dual bio-signatures of circulating tumor DNA. Biosens Bioelectron. 2015;67:443-9
163. Grieshaber D, MacKenzie R, V¨or¨os1 J, Reimhult E. Electrochemical biosensors - sensor principles and architectures. Sensors. 2008;8:1400-58
164. Pumera M, S´anchez S, Ichinose I, Tang J. Electrochemical nanobiosensors. Sens Actuator B. 2007;123:1195-1205
165. Azimzadeh M, Rahaie M, Nasirizadeh N, Ashtari K, Naderi-Manesh H. An electrochemical nanobiosensor for plasma miRNA-155,based on graphene oxide and gold nanorod, for early detection of breast cancer. Biosens Bioelectron. 2016;77:99-106
166. Rana M, Balcioglu M, Kovach M, Hizir MS, Robertson NM, Khan I, Yigit MV. Reprogrammable multiplexed detection of circulating oncomiRs using hybridization chain reaction. Chem Commun. 2016;52:3524-7
167. Guo L, Lin Y, Chen C, Qiu B, Lin Z, Chen G. Direct visualization of sub-femtomolar circulating microRNAs in serum based on the duplex-specific nuclease-amplified oriented assembly of gold nanoparticle dimers. Chem Commun. 2016;52:11347-50
168. Tatsuta M, Itoh T, Okuda S. et al. Carcinoembryonic antigen in gastric juice as an aid in diagnosis of early gastric cancer. Cancer. 1980;46:2686-92
169. Bast RCJ, Xu FJ Yu YH, Barnhill S Zhang Z. Mills GB. CA 125: the past and the future. Int J Biol Markers. 1998;13:179-87
170. Huang LJ, Chen SX, Huang Y, Luo WJ, Jiang HH, Hu QH, Zhang PF, Yi H. Proteomics-based identification of secreted protein dihydrodiol dehydrogenase as a novel serum markers of non-small cell lung cancer. Lung Cancer. 2006;54:87-94
171. Kirmiz C, Li B, An HJ. et al. A serum glycomics approach to breast cancer biomarkers. Mol Cell Proteomics. 2007;6:43-55
172. Pitteri SJ, Amon LM, Busald BT. et al. Detection of elevated plasma levels of epidermal growth factor receptor before breast cancer diagnosis among hormone therapy users. Cancer Res. 2010;70:8598-606
173. Opstal-van Winden AWJ, Krop EJM, Karedal MH, Gast MCW, Lindh CH. Searching for early breast cancer biomarkers by serum protein profiling of pre-diagnostic serum; a nested case-control study. BMC Cancer. 2011;11:381
174. Ajona D, Pajares MJ, Corrales L. Investigation of complement activation product c4d as a diagnostic and prognostic biomarker for lung cancer. J Natl Cancer Inst. 2013;105:1385-93
175. Mirabelli P, Incoronato M. Usefulness of traditional serum biomarkers for management of breast cancer patients. BioMed Res Int. 2013;2013:685641
176. Di G, Dresse M, Mayr D, Nagel D, Heinemann V, Stieber P. Serum HER2 in combination with CA15-3 as a parameter for prognosis in patients with early breast cancer. Clin Chim Acta. 2015;440:16-22
177. Fung KYC, Tabor B, Buckley MJ. et al. Blood-based proteinbiomarker panel for the detection of colorectal cancer. PLoS One. 2015;10:e0120425
178. Perkins GL, Slater ED, Sanders GK, Prichard JG. Serum tumor markers. Am Fam Physician. 2003;68:1075-82
179. Powers AD, Palecek SP. Protein analytical assays for diagnosing, monitoring, and choosing treatment for cancer patients. J Healthc Eng. 2012;3:503-34
180. Yin Y; Cao Y, Xu Y, Li G. Colorimetric immunoassay for detection of tumor markers. Int J Mol Sci. 2010;11:5077-94
181. Zhou F, Yuan L, Wang H, Li D, Chen H. Gold nanoparticle layer: a promising platform for ultra-sensitive cancer detection. Langmuir. 2011;27:2155-8
182. Wang J, Cao Y, Xu Y, Li G. Colorimetric multiplexed immunoassay for sequential detection of tumor markers. Biosens Bioelectron. 2009;25:532-6
183. Zhou WH, Zhu CL, Lu CH, Guo X, Chen F, Yang HH, Wang X. Amplified detection of protein cancer biomarkers using DNAzyme functionalized nanoprobesw. Chem Commun. 2009:6845-7
184. Liu M, Jia C, Jin Q, Lou X, Yao S, Xiang J, Zhao J. Novel colorimetric enzyme immunoassay for the detection of carcinoembryonic antigen. Talanta. 2010;81:1625-9
185. Jiang J, Zhao S, Huang Y, Qin G, Ye F. Highly sensitive immunoassay of carcinoembryonic antigen by capillary electrophoresis with gold nanoparticles amplified chemiluminescence detection. J Chromatogr A. 2013;1281:161-6
186. Hirsch LR, Jackson JB, Lee A, Halas NJ, West JL. A whole blood immunoassay using gold nanoshells. Anal Chem. 2003;75:2377-81
187. Qu W, Liu Y, Liu D, Wang Z, Jiang X. Copper-mediated amplification allows readout of immunoassays by the naked eye. Angew Chem Int Ed. 2011;50:3442-5
188. Lu C, Wang Y, Ye S, Chen G, Yang H. Ultrasensitive detection of Cu2+ with the naked eye and application in immunoassays. NPG Asia Mater. 2012;4:e10
189. Hu Y, Li L, Guo L. The sandwich-type aptasensor based on goldnanoparticles/DNA/magnetic beads for detection of cancer biomarkerprotein AGR2. Sens. Actuator B. 2015;209:846-52
190. de la Rica R, Stevens MM. Plasmonic ELISA for the ultrasensitive Detection of Disease Biomarkers with the Naked Eye. Nat Nanotechnol. 2012;7:821-4
191. Liu D, Yang J, Wang HF, Wang Z, Huang X, Wang Z, Niu G, Hight Walker AR, Chen X. Glucose oxidase-catalyzed growth of gold nanoparticles enables quantitative detection of attomolar cancer biomarkers. Anal Chem. 2014;86:5800-6
192. Bao YP, Wei TF, Lefebvre PA, An H, He L, Kunkel GT, Muller UR. Detection of protein analytes via nanoparticle-based bio bar code technology. Anal Chem. 2006;78:2055-9
193. Stoeva SI, Lee JS, Smith JE, Rosen ST, Mirkin CA. Multiplexed detection of protein cancer markers with biobarcoded nanoparticle probes. J Am Chem Soc. 2006;128:8378-9
194. Nam JM, Jang KJ, Groves JT. Detection of proteins using a colorimetric bio-barcode assay. Nat Protocols. 2007;2:1438-44
195. Thaxtona CS, Elghanian R, Thomas AD. et al. Nanoparticle-based bio-barcode assay redefines ''undetectable'' PSA and biochemical recurrence after radical prostatectomy. Proc Natl Acad Sci USA. 2009;106:18437-42
196. Kim D, Daniel WL, Mirkin CA. Microarray-based multiplexed scanometric immunoassay for protein cancer markers using gold nanoparticle probes. Anal Chem. 2009;81:9183-7
197. Tabakman SM, Lau L, Robinson JT. et al. Plasmonic substrates for multiplexed protein microarrays with femtomolar sensitivity and broad dynamic range. Nat Commun. 2011;2:466
198. Qian J, Wang C, Pan X, Liu S. A high-throughput homogeneous immunoassay based on Förster resonance energy transfer between quantum dots and gold nanoparticles. Anal Chim Acta. 2013;763:43-49
199. Nguyen HH, Park J, Kang S, Kim M. Surface plasmon resonance: a versatile technique for biosensor applications. Sensors. 2015;15:10481-510
200. Huang L, Reekmans G, Saerens D, Friedt JM, Frederix F, Francis L, Muyldermans S, Campitelli A, Hoof CV. Prostate-specific antigen immunosensing based on mixed self-assembled monolayers, camel antibodies and colloidal gold enhanced sandwich assays. Biosens Bioelectron. 2005;21:483-90
201. Choi JW, Kang DY, Jang YH, Kim HH, Mind J, Oh BK. Ultra-sensitive surface plasmon resonance based immunosensor for prostate-specific antigen using gold nanoparticle-antibody complex. Colloids Surf A. 2008;313-314:655-9
202. Sim HR, Wark AW, Lee HJ. Attomolar detection of protein biomarkers using biofunctionalized gold nanorods with surface plasmon resonance. Analyst. 2010;135:2528-32
203. Uludag Y, Tothill E. Cancer biomarker detection in serum samples using surface plasmon resonance and quartz crystal microbalance sensors with nanoparticle signal amplification. Anal Chem. 2012;84:5898-904
204. Couture M, Ray KK, Poirier-Richard HP, Crofton A, Masson JF. 96-Well plasmonic sensing with nanohole arrays. ACS Sens. 2016;1:287-94
205. Mayer KM, Hafner JH. Localized surface plasmon resonance sensors. Chem Rev. 2011;111:3828-57
206. Sriram M; Zong K, Vivekchand SRC, Gooding JJ. Single nanoparticle plasmonic sensors. Sensors. 2015;15:25774-92
207. Truong PL, Cao C, Park S, Kim M, Sim SJ. A new method for non-labeling attomolar detection of diseases based on an individual gold nanorod immunosensor. Lab Chip. 2011;11:2591-7
208. Truong PL, Kim BW, Sim SJ. Rational aspect ratio and suitable antibody coverage of gold nanorod for ultra-sensitive detection of a cancer biomarker. Lab Chip. 2012;12:1102-9
209. Rodríguez-Lorenzo L, de la Rica R, Álvarez-Puebla RA, Liz-Marzán LM, Stevens MM. Plasmonic nanosensors with inverse sensitivity by means of enzyme-guided crystal growth. Nat Mater. 2012;11:604-7
210. Lin D, Pan J, Huang H, Chen G, Qiu S, Shi H, Chen W, Yu Y, Feng S, Chen R. Label-free blood plasma test based on surface-enhanced Raman scattering for tumor stages detection in nasopharyngeal cancer. Sci Report. 2014;4:475
211. Ito H, Inoue H, Hasegawa K, Hasegawa Y, Shimizu T, Kimura S, Onimaru M, Ikeda H, Kudo S. Use of surface-enhanced Raman scattering for detection of cancer-related serum-constituents in gastrointestinal cancer patients. Nanomedicine. 2014;10:599-608
212. Lv Y, Qin Y, Svec F, Tan T. Molecularly imprinted plasmonic nanosensor for selective SERS detection of protein biomarkers. Biosens Bioelectron. 2016;80:433-41
213. Grubisha DS, Lipert RJ, Park HY, Driskell J, Porter MD. Femtomolar detection of prostate-specific antigen: an immunoassay based on surface-enhanced Raman scattering and immunogold labels. Anal Chem. 2003;75:5936-43
214. Li M, Kang JW, Sukumar S, Dasari RR, Barman I. Multiplexed detection of serological cancer markers with plasmon-enhanced Raman spectroimmunoassay. Chem Sci. 2015;6:3906-14
215. Li M, Cushing SK, Zhang J, Suri S, Evans R, Petros WP, Gibson LF, Ma D, Liu Y, Wu N. Three-dimensional hierarchical plasmonic nano-architecture enhanced surface-enhanced Raman scattering immunosensor for cancer biomarker detection in blood plasma. ACS Nano. 2013;7:4967-76
216. Krasnoslobodtsev AV, Torres MP, Kaur S, Vlassiouk IV, Lipert RJ, Jain M, Batra SK, Lyubchenko YL. Nano-immunoassay with improved performance for detection of cancer biomarkers. Nanomedicine. 2015;11:167-73
217. Chon H, Lee S, Son SW, Oh CH, Choo J. Highly sensitive immunoassay of lung cancer marker carcinoembryonic antigen using surface-enhanced Raman scattering of hollow gold nanospheres. Anal Chem. 2009;81:3029-34
218. Chon H, Lee S, Yoon SY, Chang SI, Lim DW, Choo J. Simultaneous immunoassay for the detection of two lung cancer markers using functionalized SERS nanoprobes. Chem Commun. 2011;47:12515-7
219. Wang G, Lipert RJ. Detection of the potential pancreatic cancer marker MUC4 in serum using surface-enhanced Raman scattering. Anal Chem. 2011;83:2554-61
220. Li J, Skeete Z, Shan S, Yan S, Kurzatkowska K, Zhao W, Ngo QM, Holubovska P, Luo J, Hepel M, Zhong CJ. Surface enhanced Raman scattering detection of cancer biomarkers with bifunctional banocomposite probes. Anal Chem. 2015;87:10698-702
221. Lin G, Feng S, Pan J, Chen Y, Lin J, Chen G, Xie S, Zeng H, Chen R. Colorectal cancer detection by gold nanoparticle based surface-enhanced Raman spectroscopy of blood serum and statistical analysis. Optic Express. 2011;19:13565-77
222. Lin D, Pan J, Huang H, Chen G, Qiu S, Shi H, Chen W, Yu Y, Feng S, Chen R. Label-free blood plasma test based on surface-enhanced Raman scattering for tumor stages detection in nasopharyngeal cancer. Sci Report. 2014;4:srep04751
223. Feng S, Chen R, Lin J, Pan J, Chen G, Li Y, Cheng M, Huang Z, Chen J, Zeng H. Nasopharyngeal cancer detection based on blood plasma surface-enhanced Raman spectroscopy and multivariate analysis. Biosens Bioelectron. 2010;25:2414-9
224. Feng S, Chen R, Lin J, Pan J, Wu Y, Li Y, Chen J, Zeng H. Gastric cancer detection based on blood plasma surface-enhanced Raman spectroscopy excited by polarized laser light. Biosens Bioelectron. 2011;26:3167-74
225. Feng S, Wang W, Tai IT, Chen G, Chen R, Zeng H. Label-free surface-enhanced Raman spectroscopy for detection of colorectal cancer and precursor lesions using blood plasma. BioMed Opt Express. 2015;6:3494-502
226. Ravalli A, dos Santos GP, Ferroni M, Faglia G, Yamanaka H, Marrazza G. New label free CA125 detection based on gold nanostructured screen-printedelectrode. Sensors Actuators B: Chem. 2013;179:194-200
227. Raghav R, Srivastava S. Core-shell gold-silver nanoparticles based impedimetric immunosensor for cancer antigen CA125. Sens Actuator B. 2015;220:557-64
228. Arkan E, Saber R, Karimi Z, Shamsipur M. A novel antibody-antigen based impedimetric immunosensor for low level detection of HER2 in serum samples of breast cancer patients via modification of a gold nanoparticles decorated multiwall carbon nanotube-ionic liquid electrode. Anal Chim Acta. 2015;874:66-74
229. Feng D, Li L.; Zhao J, Zhang Y. Simultaneous electrochemical detection of multiple biomarkers using gold nanoparticles decorated multiwall carbon nanotubes as signalenhancers. Anal Biochem. 2015;482:48-54
230. Presnova G, Presnov D, Krupenin V, Grigorenko V, Trifonov A, Andreeva I, Ignatenko O, Egorov A, Rubtsova M. Biosensor based on a silicon nanowire field-effect transistor functionalized by gold nanoparticles for the highly sensitive determination of prostate specific antigen. Biosens Bioelectron. 2016
231. Ilkhani H, Sarparast M, Noori A, Bathaie SZ, Mousavi MF. Electrochemical aptamer/antibody based sandwich immunosensor for the detection of EGFR, a cancer biomarker using gold nanoparticlesas a signaling probe. Biosens Bioelectron. 2015;74:491-197
232. Liu X, Qin Y, Deng C, Xiang J, Li Y. A simple and sensitive impedimetric aptasensor for the detection of tumor markers based on gold nanoparticles signal amplification. Talanta. 2015;132:150-54
233. Liu X, Dai Q, Austin L, Coutts J, Knowles G, Zou J, Chen H, Huo Q. A one-step homogeneous immunoassay for cancer biomarker detection using gold nanoparticle probes coupled with dynamic light scattering. J Am Chem Soc. 2008;130:2780-2
234. Zheng T, Pierre-Pierre N, Yan X. et al. Gold nanoparticle-enabled blood test for early stage cancer detection and risk assessment. ACS Appl Mater Interfaces. 2015;7:6819-27
235. Kosaka PM, Pini V, Ruz JJ, da Silva RA, González MU, Ramos D, Calleja M, Tamayo J. Detection of cancer biomarkers in serum using a hybrid mechanical and optoplasmonic nanosensor. Nat Nanotechnol. 2014;9:1047-53
236. Zavaleta CL, Smith BR, Walton I, Doering W, Davis G, Shojaei B, Natan MJ, Gambhir SS. Multiplexed imaging of surface enhanced Raman scattering nanotags in living mice using noninvasive Raman spectroscopy. Proc Nat Aca Sci USA. 2009;106:13511-66
237. Mao X, Gurung A, Xu H, Baloda M, He Y, Liu G. Simultaneous detection of nucleic acid and protein using gold nanoparticles and lateral flow device. Anal Sci. 2014;30:637-42
Author Biography
 Dr. Xiaohua Huang is an associate professor of chemistry and biomedical engineering at The University of Memphis, USA. She received her Ph.D. degree in chemistry from Georgia Institute of Technology in 2006 and postdoctoral training at Georgia Institute of Technology and Emory University from 2006 to 2010. She has published 50 peer- reviewed journal articles and 7 book chapters. The current research interests in Dr. Huang's group include: (1) Shape-controlled synthesis and properties of magnetic-plasmonic core-shell nanoparticles; (2) Single particle optical imaging and spectroscopy; (3) Isolation, detection, and analysis of circulating tumor cells; and (4) Detection and analysis of exosomes and microvesicles.
Dr. Xiaohua Huang is an associate professor of chemistry and biomedical engineering at The University of Memphis, USA. She received her Ph.D. degree in chemistry from Georgia Institute of Technology in 2006 and postdoctoral training at Georgia Institute of Technology and Emory University from 2006 to 2010. She has published 50 peer- reviewed journal articles and 7 book chapters. The current research interests in Dr. Huang's group include: (1) Shape-controlled synthesis and properties of magnetic-plasmonic core-shell nanoparticles; (2) Single particle optical imaging and spectroscopy; (3) Isolation, detection, and analysis of circulating tumor cells; and (4) Detection and analysis of exosomes and microvesicles.
 Ryan O'Connor received his bachelor's degree in chemistry from Xavier University in 2013. He is currently a Ph.D. student under the supervision of Dr. Xiaohua Huang. His research is to improve the detection and analysis of circulating tumor cells through the use of surface enhanced Raman scattering nanoparticles and microfluidics platform.
Ryan O'Connor received his bachelor's degree in chemistry from Xavier University in 2013. He is currently a Ph.D. student under the supervision of Dr. Xiaohua Huang. His research is to improve the detection and analysis of circulating tumor cells through the use of surface enhanced Raman scattering nanoparticles and microfluidics platform.
 Elyahb Allie Kwizera received his bachelor's degree in chemistry and mathematics from the University of Central Arkansas in 2014. He is currently a Ph.D. student under the supervision of Dr. Xiaohua Huang. His research is centered on controlled synthesis, properties and applications of magnetic-plasmonic core-shell nanoparticles as well as development of novel nanotechnology-based technologies for detection and analysis of circulating vesicles.
Elyahb Allie Kwizera received his bachelor's degree in chemistry and mathematics from the University of Central Arkansas in 2014. He is currently a Ph.D. student under the supervision of Dr. Xiaohua Huang. His research is centered on controlled synthesis, properties and applications of magnetic-plasmonic core-shell nanoparticles as well as development of novel nanotechnology-based technologies for detection and analysis of circulating vesicles.
![]() Corresponding author: Email: xhuang4edu. Phone: (901) 678 1728. Fax: (901) 678 3744.
Corresponding author: Email: xhuang4edu. Phone: (901) 678 1728. Fax: (901) 678 3744.

 Global reach, higher impact
Global reach, higher impact